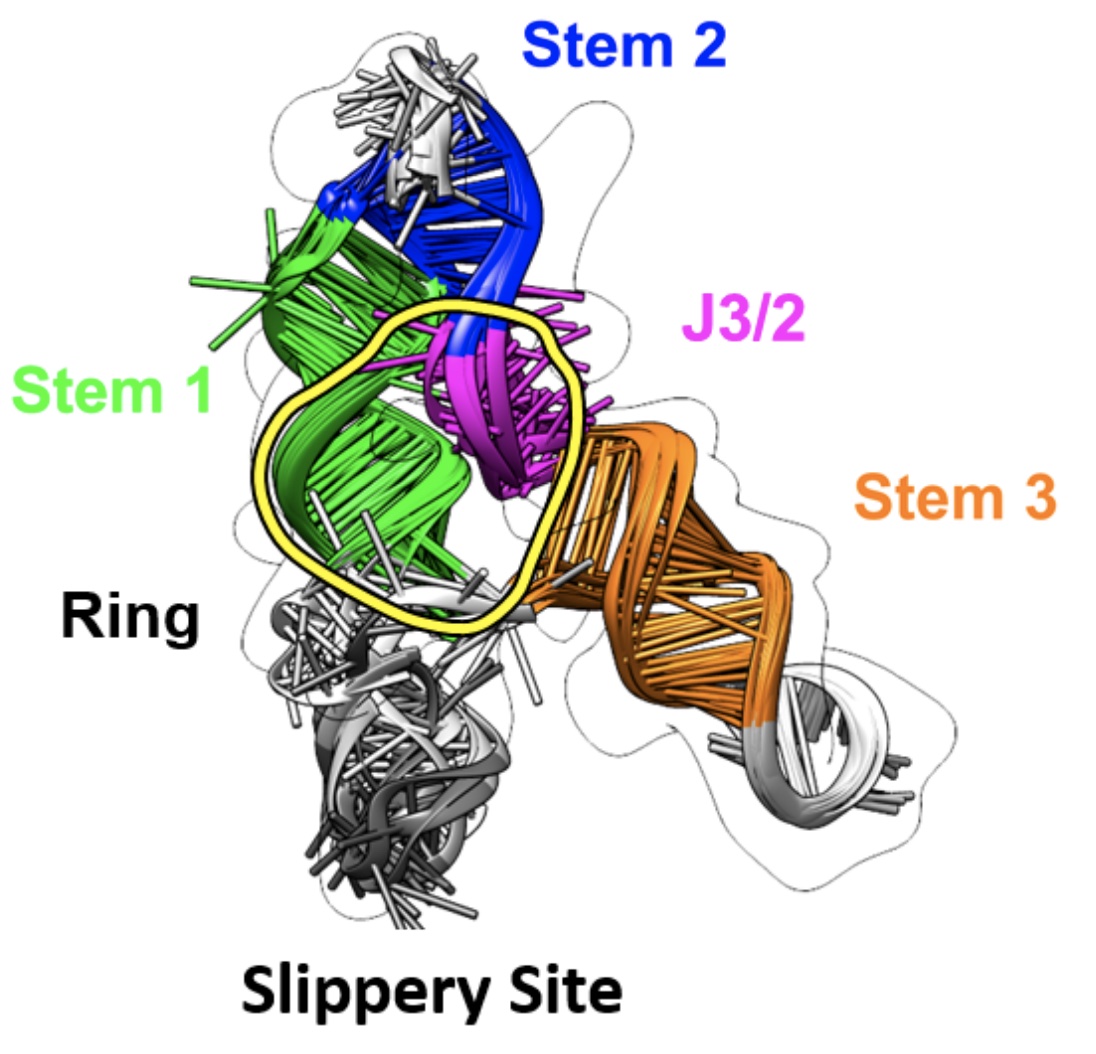
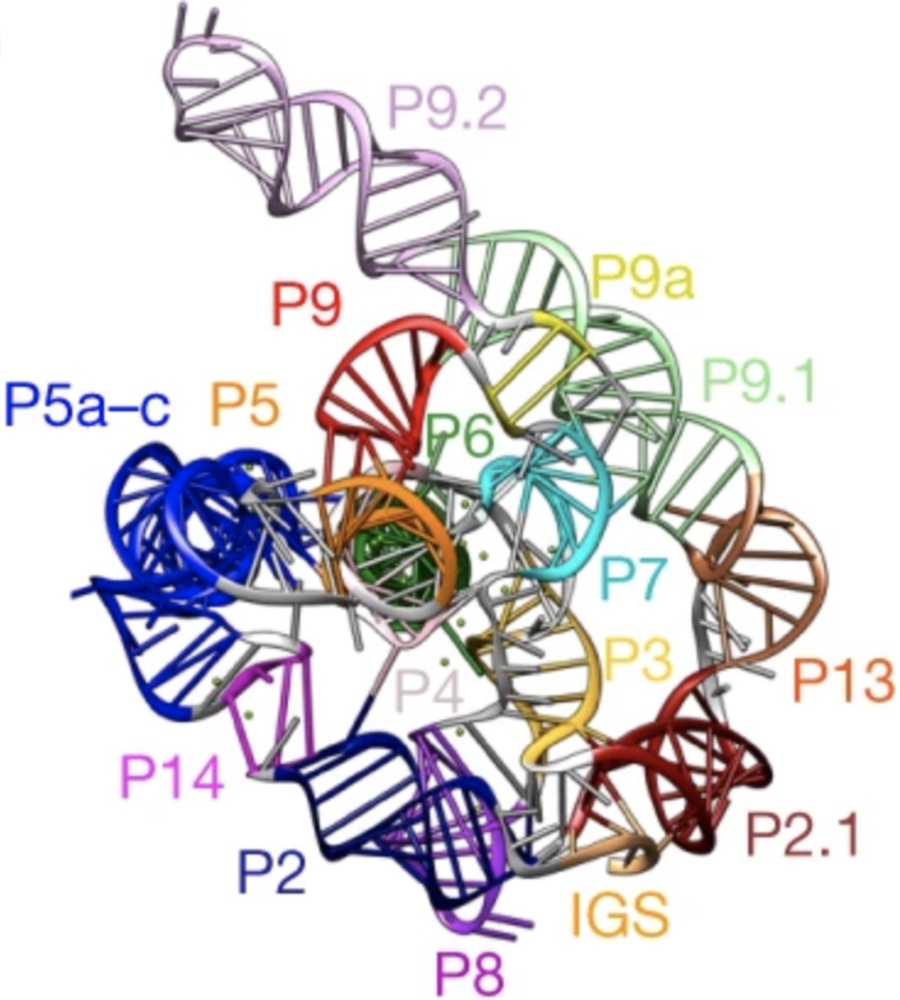
Congratulations to Alexis Van Zandt for being named an HBCU STEM queen! She’s on the cover of Ebony magazine! Alexis, a recent graduate of Prairie View A&M University, worked with the Das laboratory and the Center for Genetically Engineered Materials to develop a better design-oriented understanding of the ribosome in 2021.
Congratulations to Alexis Van Zandt for being named an HBCU STEM queen! She’s on the cover of Ebony magazine! Alexis, a recent graduate of Prairie View A&M University, worked with the Das laboratory and the Center for Genetically Engineered Materials to develop a better design-oriented understanding of the ribosome in 2021.