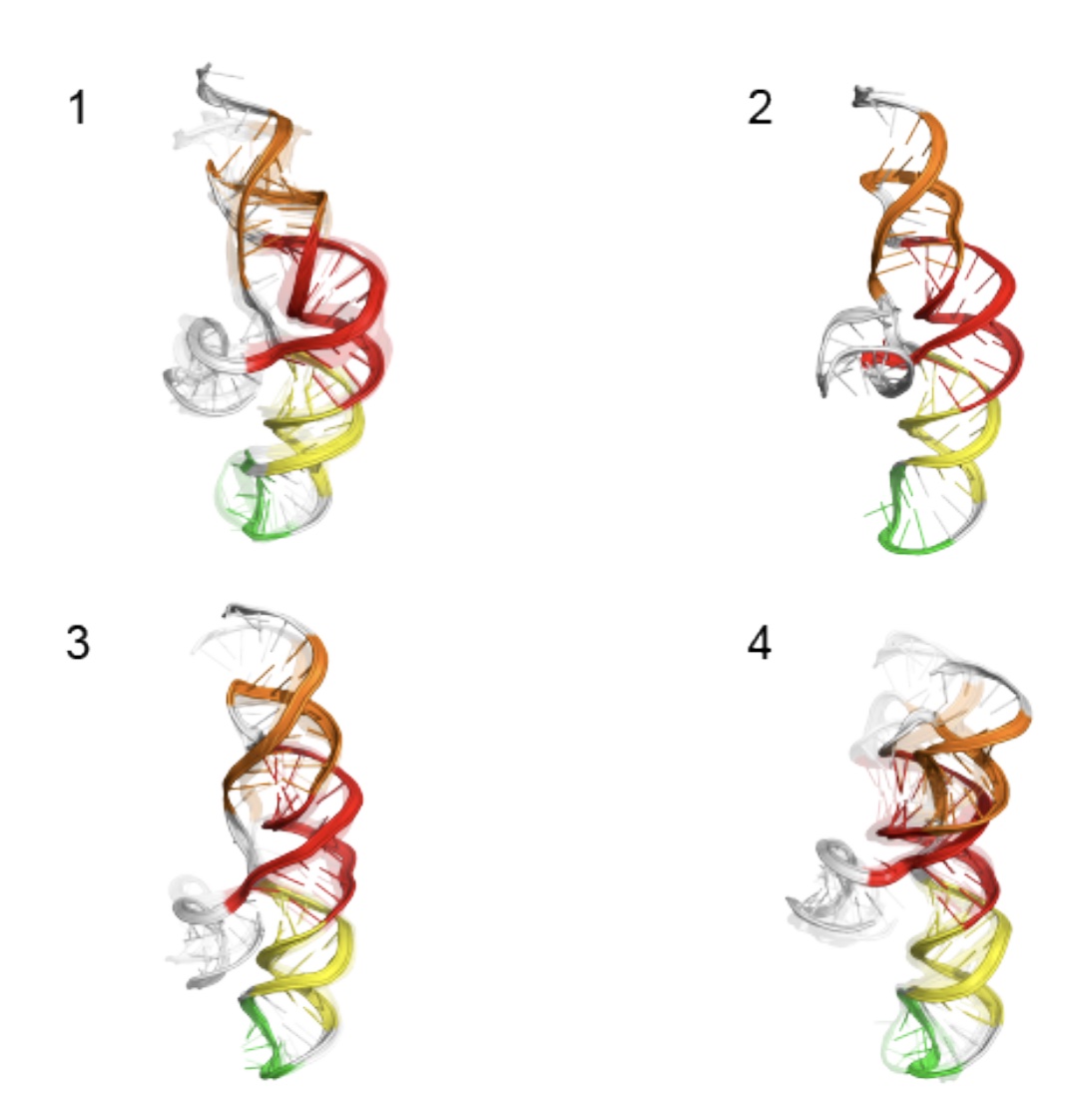
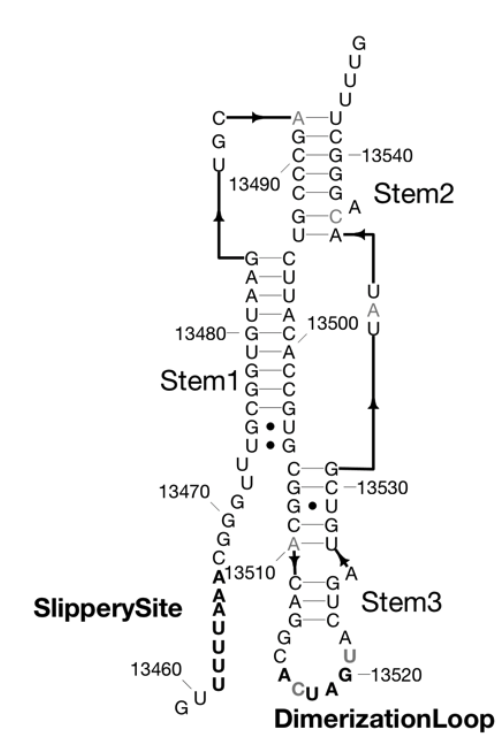
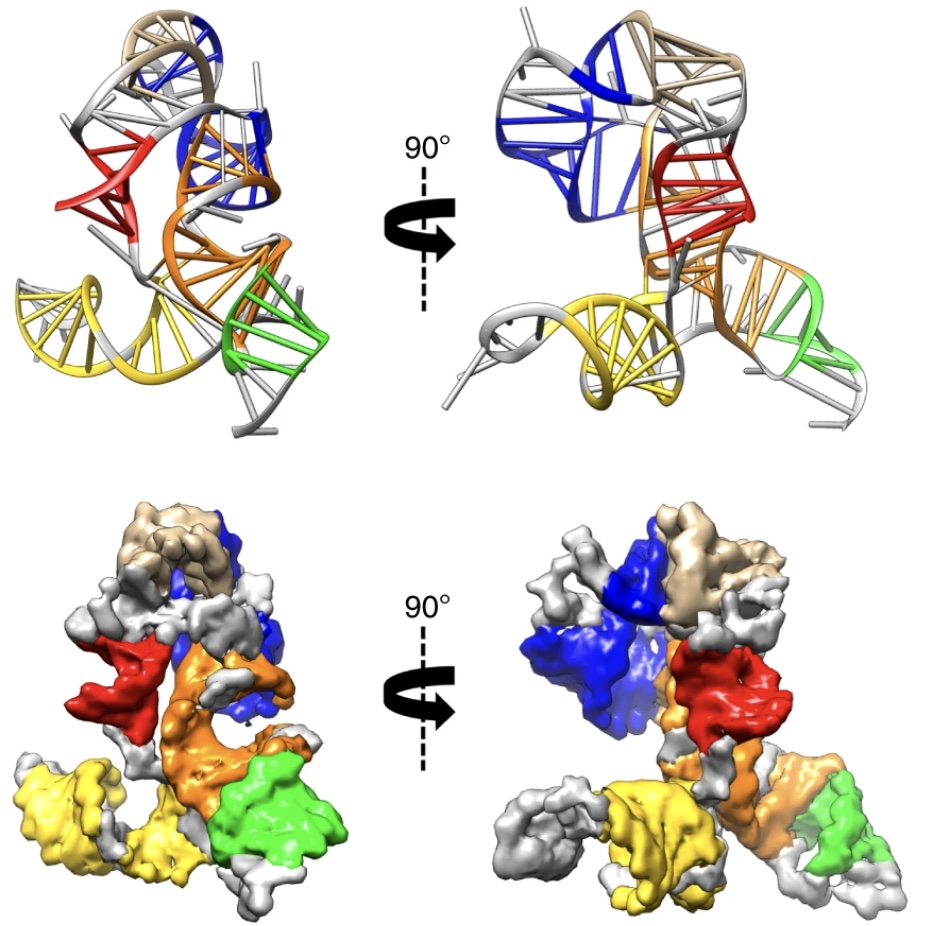
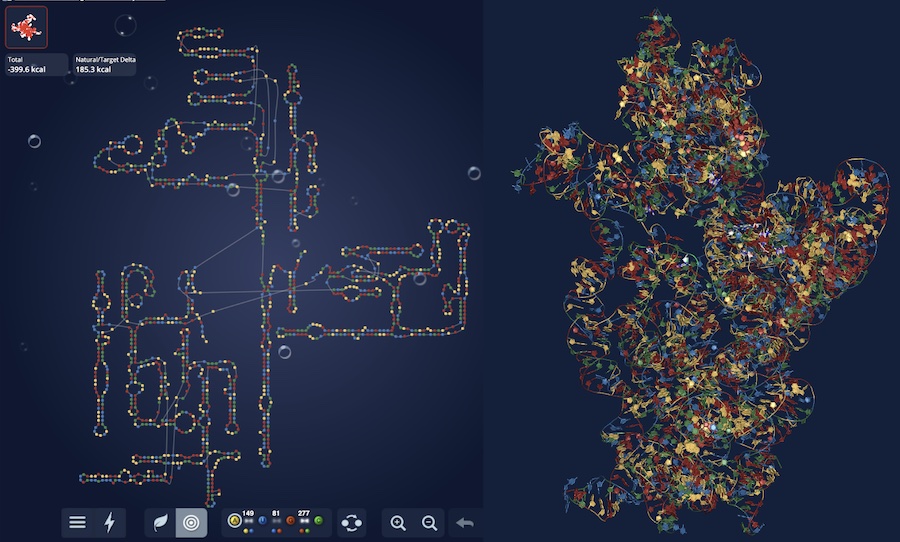
There are many conserved and likely structured elements of the SARS-CoV-2 genome, such as those identified in a previous preprint, with no known 3D structure, which nonetheless may be useful targets for small molecule therapeutics. Ramya Rangan and Andy Watkins have performed studies with our latest structure prediction code, FARFAR2, to make publicly available collections of de novo 3D models of these elements. Their linked preprint has been made available in bioRxiv.
There are many conserved and likely structured elements of the SARS-CoV-2 genome, such as those identified in a previous preprint, with no known 3D structure, which nonetheless may be useful targets for small molecule therapeutics. Ramya Rangan and Andy Watkins have performed studies with our latest structure prediction code, FARFAR2, to make publicly available collections of […]