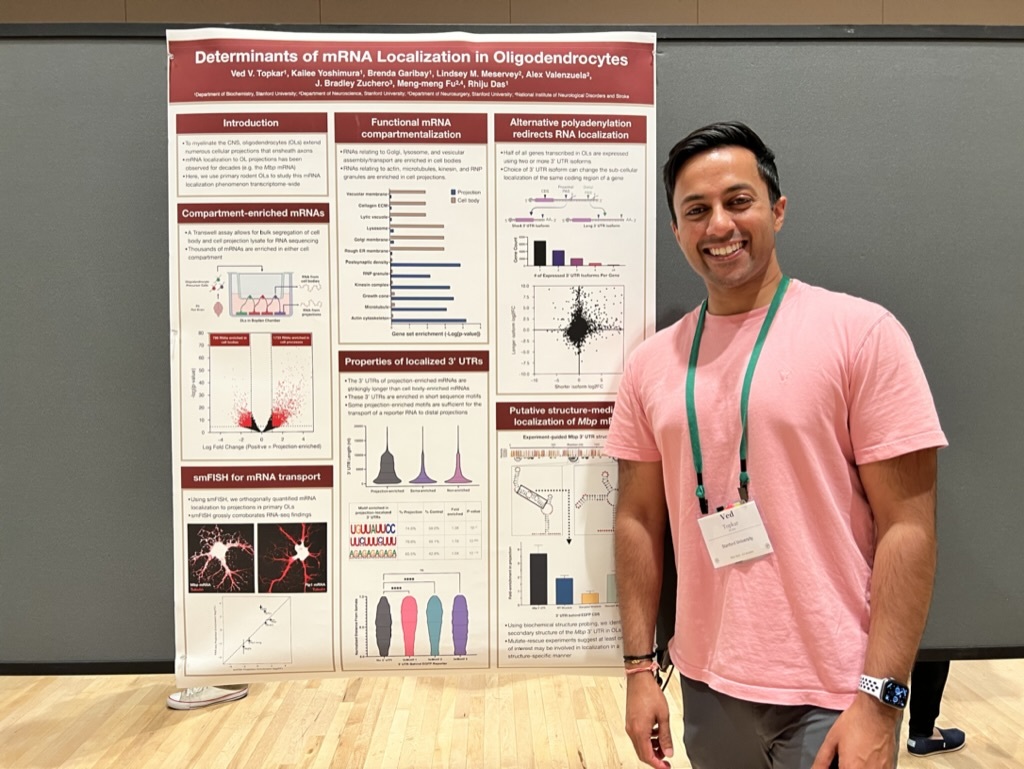
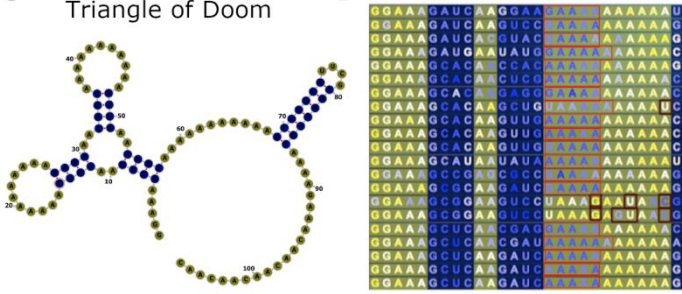
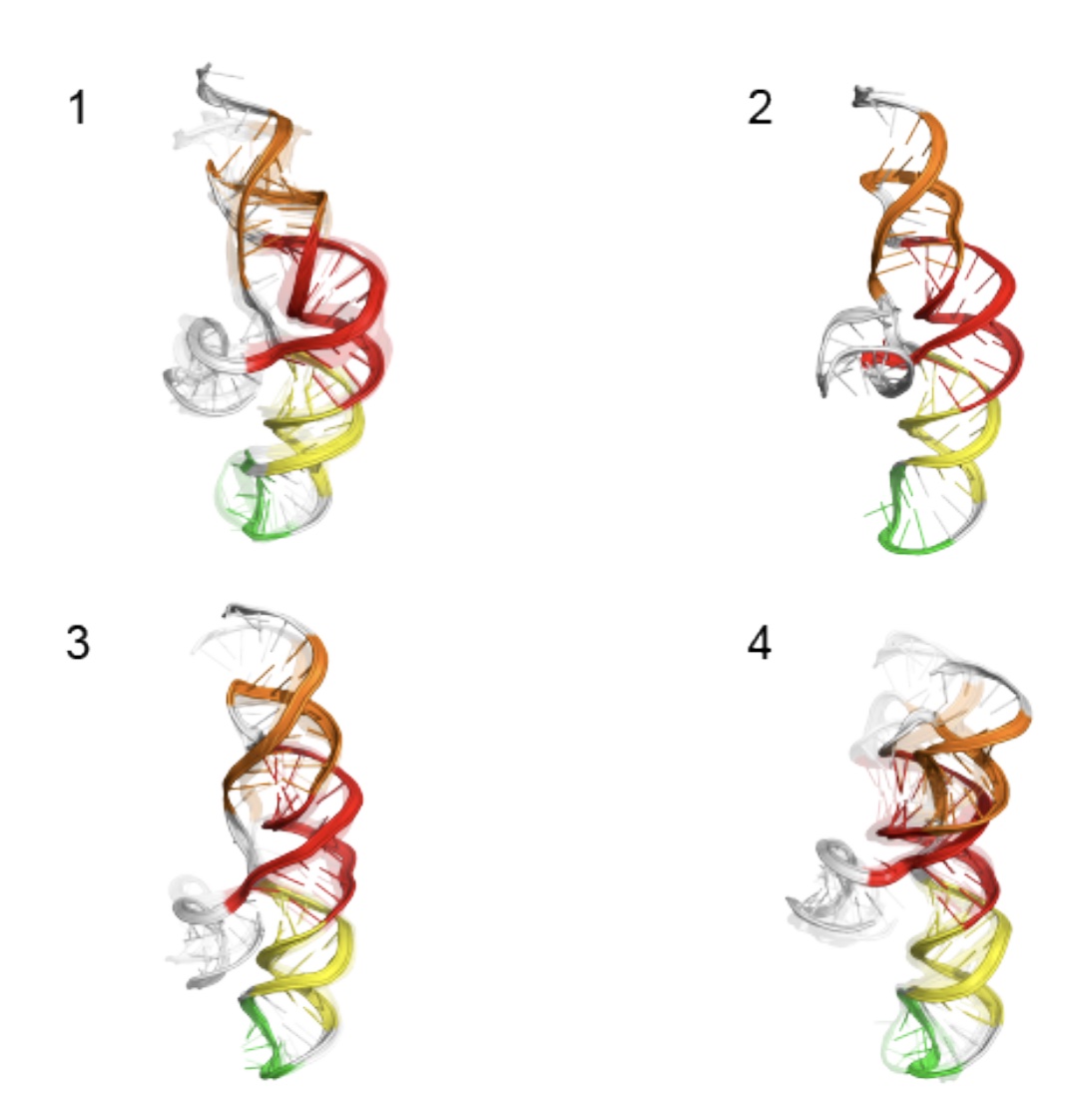
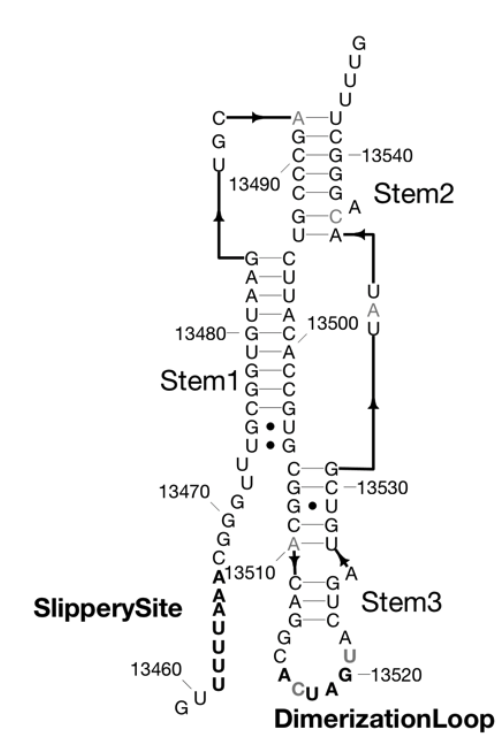
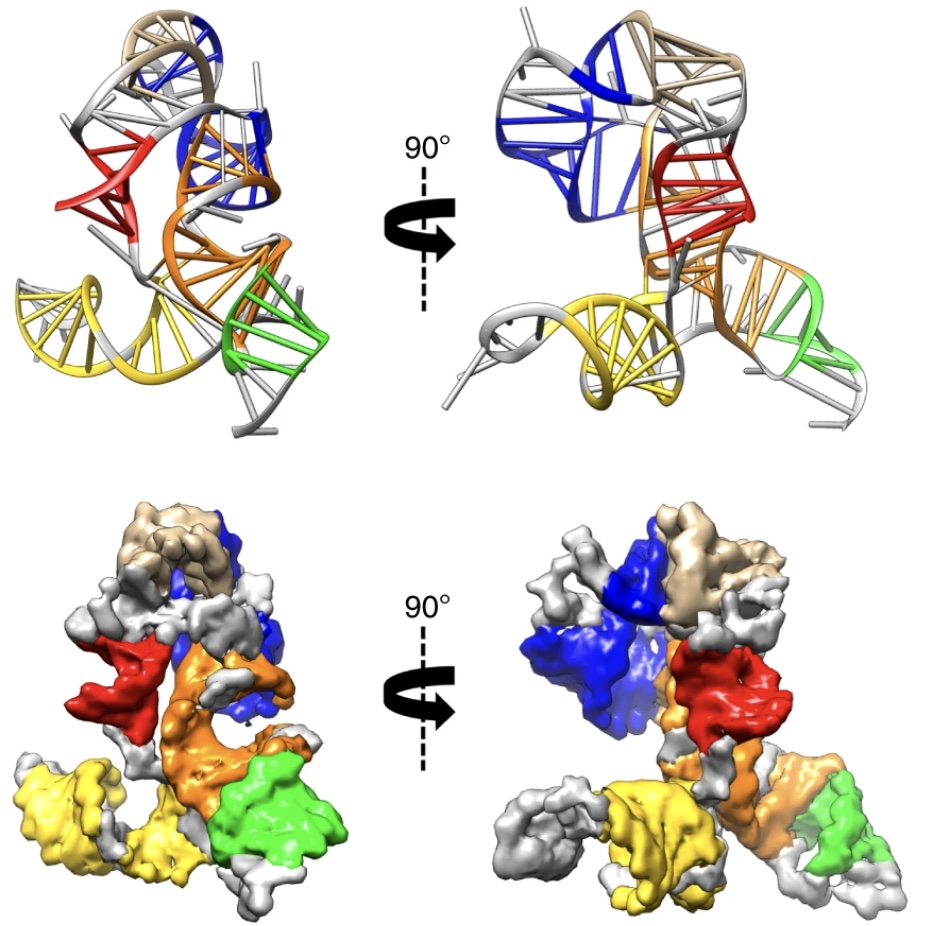
Congrats to Ved Topkar for succesfully defending his thesis on “Determinants of mRNA Localization in Oligodendrocytes”!
We’re hiring again! The Das lab is looking for a Life Sciences Technician II to help us acquire RNA structural data on 1 billion diverse molecules! If you’re interested in doing in-depth research involving high throughput biochemistry and structure mapping, check out the job posting and apply here!
Rhiju talked about “Wordwide Competitions in the RNA Folding Problem” at the Bio ML seminar series at U.C. Berkeley. Check out the full schedule at the BioML website and the talk is on YouTube.
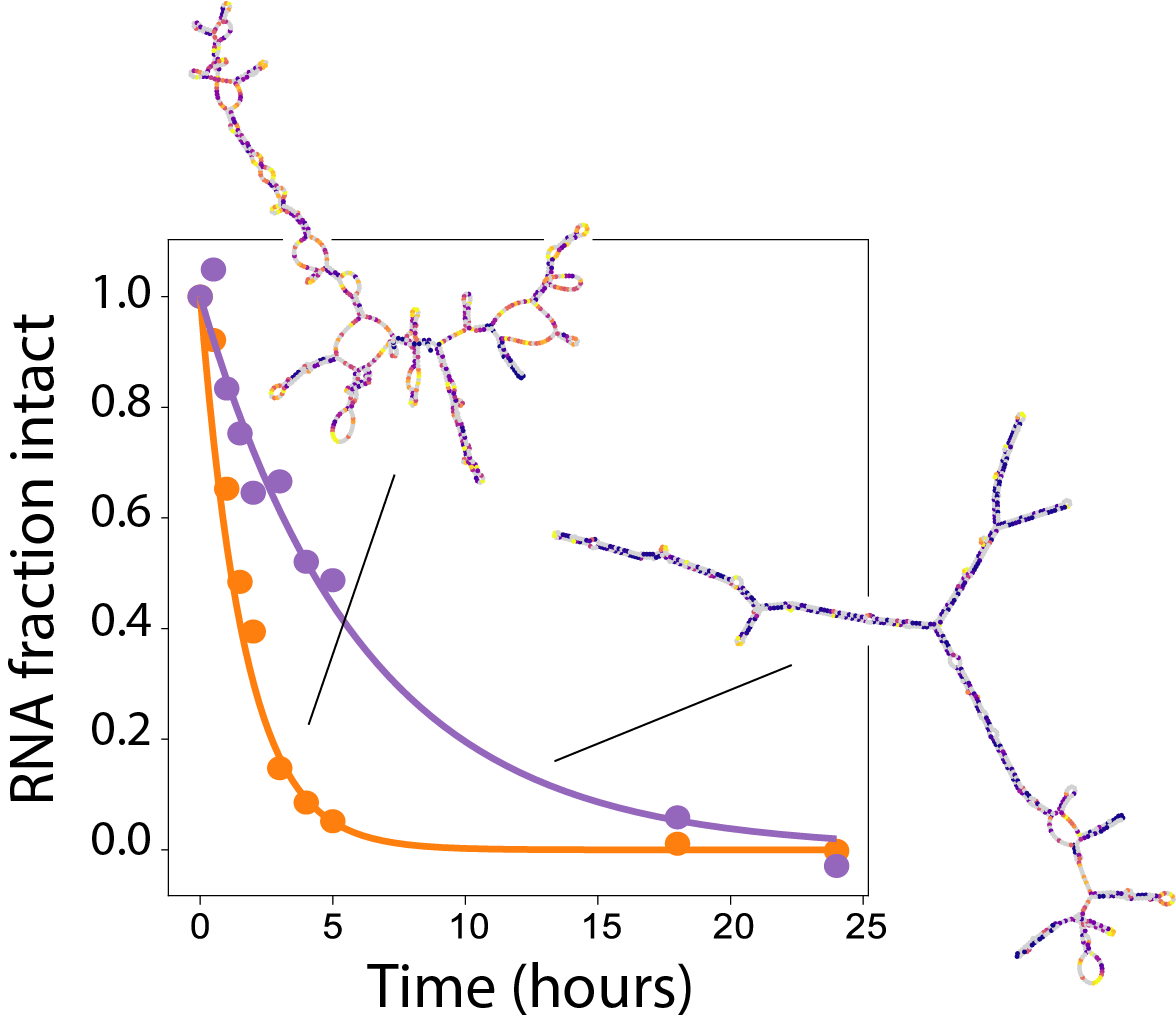
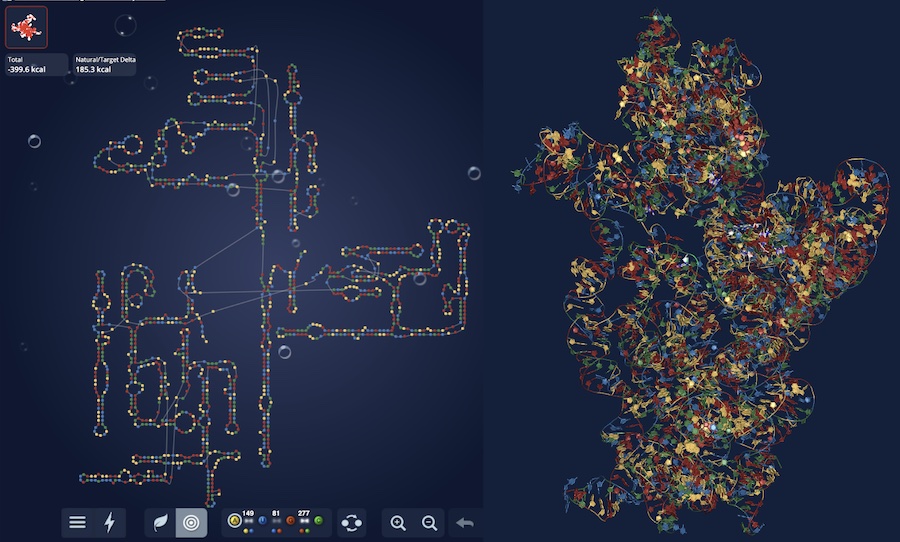
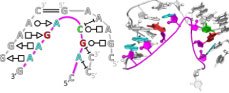
We’ve released chemical mapping data on 2.1M RNA sequences as well as a deep learning model RibonanzaNet, distilled from an 800-team Kaggle competition and the Eterna OpenKnot challenge. All data and models are publicly available — check out our preprint: Ribonanza: deep learning of RNA structure through dual crowdsourcing.
We’ve launched an online Stanford Coursera course: RNA Biology with Eterna. Learn the basics of RNA biology from the point of view of the molecules’ folding and biochemistry. It’s a totally free course, targeting students at college level, with interactive components based on the Eterna videogame.
Call to action to RNA and ML folks everywhere: We are getting ready to scale up our chemical mapping data protocol, initially to 8M sequences, and then onwards to 100 M and, hopefully 1B, sequences. For this Ribonanza 2 initiative, we’re looking for diverse sets of RNA molecules from as many different groups as possible! Do you have a list of natural or designed RNA sequences that you’re curious to visualize? We collected the first round in January 15, 2024, and if you’re interested in joining the next round, e-mail us at ribonanza [at] stanford.edu.
Rhiju spoke at the MLSB workshop at NeurIPS on Dec. 15, 2023, in New Orleans. We announced results of the Ribonanza competition. Impressive models came in from many teams demonstrating the deep learning of RNA structure from a data set involving 2.1M RNA sequences. But it’s not over yet — different models predict different answers for new RNAs — stay tuned for Ribonanza 2!
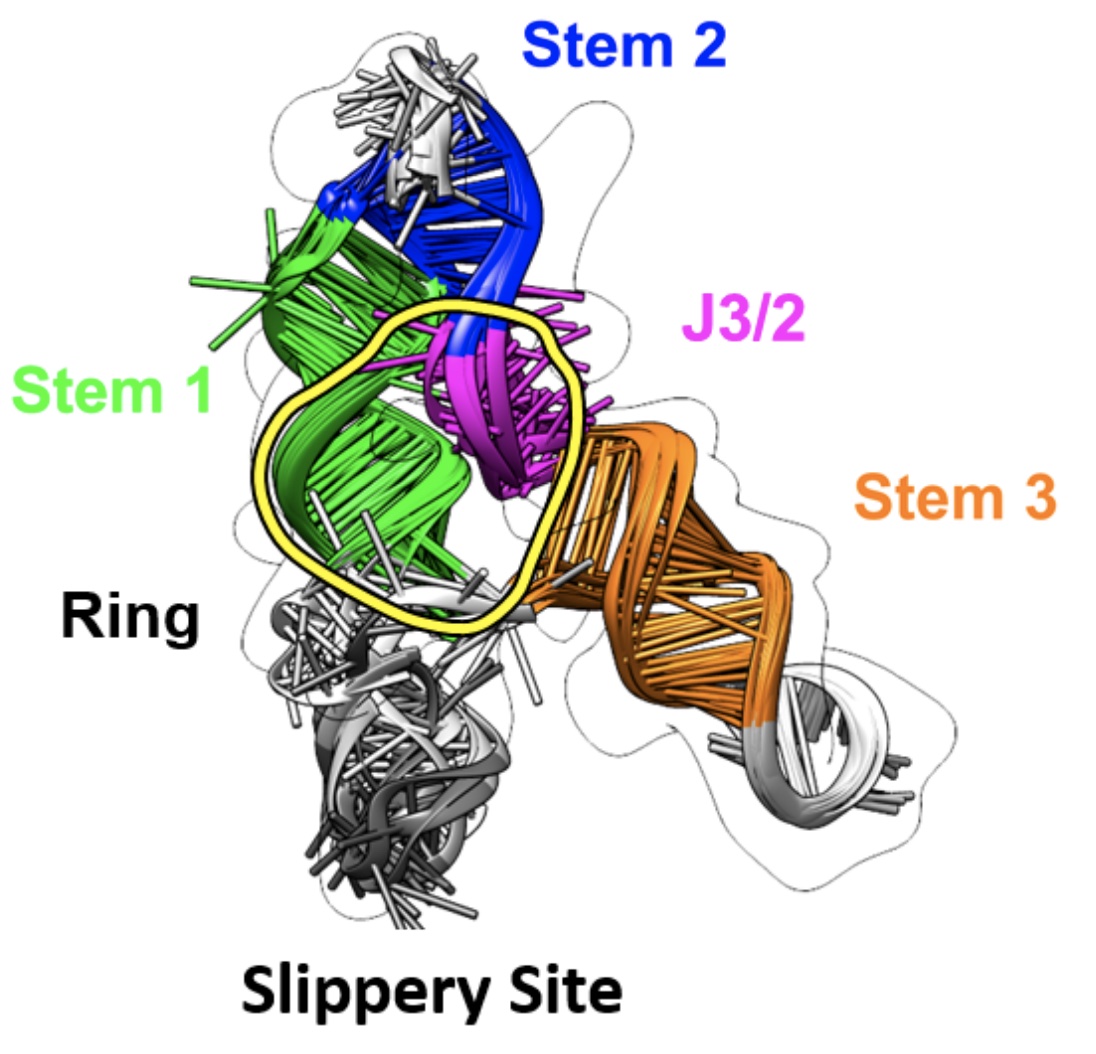
It was exciting to meet the next generation of scientists at ABRCMS in Phoenix! Congrats to lab postbacc Josh Johnson, who won a poster award for his work on Coronavirus Frameshift Stimulating Element Structure Determination and Therapeutic Design. Josh is pictured here with Rhiju and Gloria Regisford, Josh’s undergraduate mentor at Prairie View A&M. Also wonderful to coordinate with Jill Townley from Eterna, who demo-ed research and education on Eterna at the ABRCMS RNA Society booth!
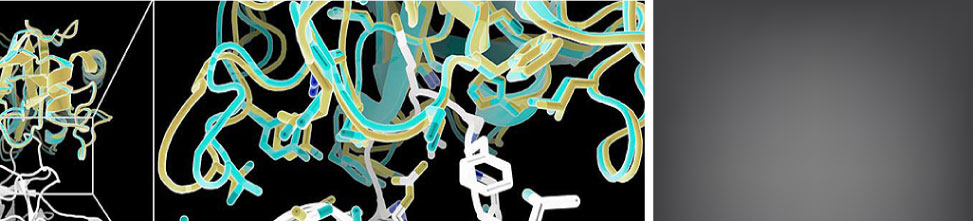
The version of record of our CASP15 RNA assessment paper is out in the Proteins special issue. Read about it here. Also in the same issue: a perspective from groups (including ours) who experimentally solved the CASP15 3D RNA structures, as well as papers from the four top predictor groups, AIchemy_RNA2, Chen, RNAPolis, and GeneSilico. None used deep learning!
Rhiju gave an online talk on Ribonanza: big data for RNA structure prediction at the CASP RNA special interest group. The talk is now available on YouTube, and previous and upcoming seminars are available on the RNA SIG channel!
We’ve teamed up with Kaggle to accelerate progress in RNA structure prediction through a new machine learning competition. Check out the new Ribonanza challenge! Register by Nov. 30, 2023, deadline for model submission is Dec. 7, 2023. Winning models to be open sourced and presented at NeurIPS MLSB workshop. Visit the competition webpage.
Congrats to Ana Espeleta, RosettaCommons REU, for presenting her work on “Training an RNA sequence designer using deep learning neural networks” at RosettaCon 20!
Rhiju was honored to join many of his heroes in RNA science at the RNA Worlds symposium at U. Colorado, Boulder. Talks from his session, The Future World of RNA are now available on YouTube!
Ramya Rangan presented her work on the RNA structure landscape of yeast introns at the RNA society 2023 meeting in Singapore!
Rhiju presented a short talk on Progress in RNA 3D modeling? at the RNA collaborative seminar series, along with Gloria Brar (U.C. Berkeley) speaking on unexpected RNAs and off-target effects from genomic expression casettes. You can check out our talks here. Thanks to Bay Area RNA Club for choosing us as their representatives, and to the RNA society for hosting these seminars. And our bioRxiv preprint on CASP15 RNA assessment is also out here!
Congratulations to Jose Chacon, Ph.D. student at UCSD who worked with the Das lab in 2020-21 as a SSRP student-turned-research-affiliate — Jose has just been awarded the NSF Graduate Research Fellowship!
Congratulations to Ramya Rangan for successfully defending her Ph.D. thesis in Biophysics! Ramya’s thesis has focused on RNA structures in non-coding RNA, from the SARS-CoV-2 genome to yeast introns!
Ernst Pulido, current lab rotation student, spoke at the UCSF Gladstone Institute on his career pathway as a first-generation scholar, at an event announcing Gladstone’s expansion with a visit from San Francisco Mayor London Breed!
Rhiju spoke at the The RNA World session as part of the 67th Biophysical Society annual meeting on Feb. 19, 2023.
Rachael Kretsch, Phillip Pham, Ramya Ranga, and Rhiju assessed the RNA category at CASP15! It was a lot of fun to co-assess with the RNA-puzzles team and to work with the CASP organizers. This picture is from the conference in Antalya, Turkey and shows Rachael, Rhiju, John Moult (CASP), and Eric Westhof (RNA-Puzzles). Learn more about the assessment at the CASP15 website and our presentation. And the talk recording is now up on YouTube too.
Rhiju spoke on “RNA modelling and design” at the Next Generation Biophysics Symposium 2022 hosted by the MRC Laboratory of Molecular Biology in the UK. The focus was on cryo-EM to solve RNA 3D structures!
Welcome Vivian Wu and Rui Huang to the Das lab! Vivian is joining as the Das lab manager and Rui is joining as a lab tech.
We’re hiring a Lab Manager! If you’re interested in managing a small to mid-sized lab who loves RNA structure/function and cares deeply about diversity in the scientific community, please apply to our HHMI-based job here!
We’re hiring again! The Das lab is looking for a Life Sciences Technician to work on structures involved in RNA splicing in eukaryotes! If you’re interested in doing in-depth research involving high throughput biochemistry and structure mapping, check out the job posting and apply here! This is a second job posting — we’re also looking for candidates to study structures involved in RNA transport in the nervous system here.
We’re hiring! The Das lab is looking for a Life Sciences Technician to work on detection and functional probing of RNA structures in the nervous system. It’s a two year project involving close mentorship from our lab members in the molecular biology, cell culture, microscopy, and sequencing needed to complete some fundamental papers in how RNA works in neurobiology and disease. If you’re interested in doing in-depth research, check out the job posting and apply here!
We were thrilled that the majority of the lab was able to attend the RNA 2022 conferencein Boulder, Colorado! Congrats to Ved Topkar for winning a Poster Award for his poster deciphering RNA Transport in oligodendrocytes!
We were thrilled that the majority of the lab was able to attend the RNA 2022 conferencein Boulder, Colorado! Congrats to Ved Topkar for winning a Poster Award for his poster deciphering RNA Transport in oligodendrocytes!
We’re proud to be part of one of 12 centers selected as an Antiviral Drug Discovery (AViDD) Center for Pathogens of Pandemic Concern. We look forward to continuing the work we did in collaboration with the labs of Jeff Glenn, Wah Chiu, Ralph Baric, and other colleagues in SARS-CoV-2 to develop Outpatient Antiviral Cocktails against SARS-CoV-2 and other Potential Pandemic RNA Viruses! Read more at the NIH website.
Rachael Kretsch presented the SLAC Public Lecture on Thursday, April 7, 2022 on a very exciting topic: “Revolutionary 3-D Views of Viral RNA Using Cryogenic Electron Microscopy”. Come join us by Zoom!
Congratulations to Alexis Van Zandt for being named an HBCU STEM queen! She’s on the cover of Ebony magazine! Alexis, a recent graduate of Prairie View A&M University, worked with the Das laboratory and the Center for Genetically Engineered Materials to develop a better design-oriented understanding of the ribosome in 2021.
We’re tremendously excited and honored at Rhiju’s selection in the most recent cohort of amazing HHMI Investigators. Big thanks to current lab members, lab alumni, the Eterna project and many wonderful collaborators in the RNA world for making this possible!
Rosetta REU researcher Philip Pham joined our lab to test the ARES neural network for cryo-EM — we’re delighted that Philip will continue with us through the next academic year.
Congrats to Grace Nye, Brenda Garibay, and Kailee Yoshimura for their awesome research on viral RNA structure, 3′ UTR isoforms, and smiFISH probes in our lab, as part of the SSRP program. Special congratulations to Brenda and Kailee who took home awards for best presentations at the SSRP symposium!
We’ve been delighted to host Alexis Van Zandt and Dina Tekle, both from Prairie View A&M, for the CGEM REU program this summer. Both Alexis and Dina were part of ‘Team ribosome’ and worked to develop a database of ribosome mutations and their functions.
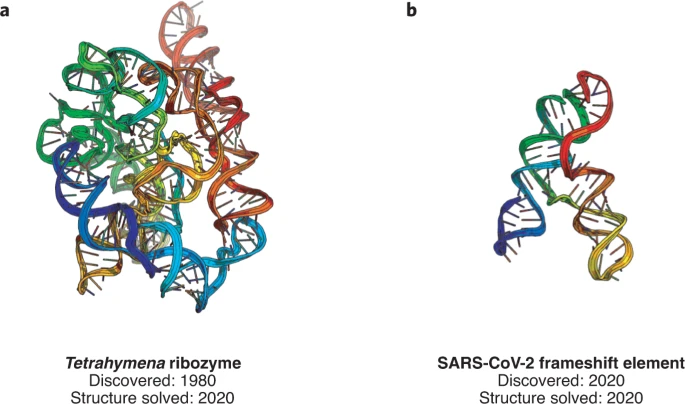
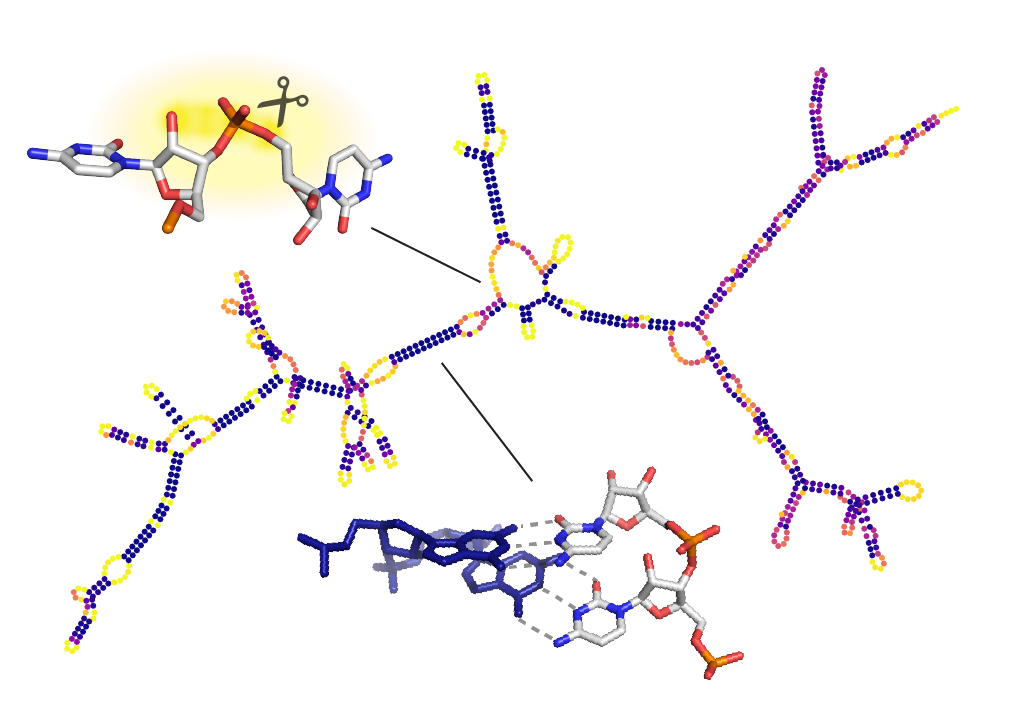
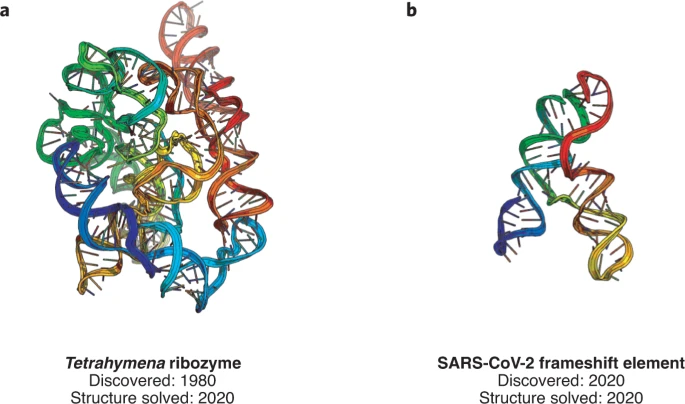
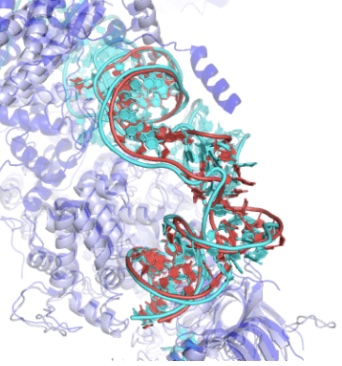
We’re excited to publish our cryo-EM structure of the frameshift stimulation element of the SARS-CoV-2 RNA genome in Nature Structure and Molecular Biology. This work appeared on bioRxiv in 2020, and the new paper includes improved antisense oligonucleotide that target this intricate RNA fold. Thanks to many labs, including those of Jeff Glenn, Wah Chiu, Tim Sheahan, Ralph Baric, and Victoria D’Souza, for this collaboration! You can read more about our work to rapidly solve RNA structures with Wah Chiu at this SLAC piece that also includes the Tetrahymena ribozyme.
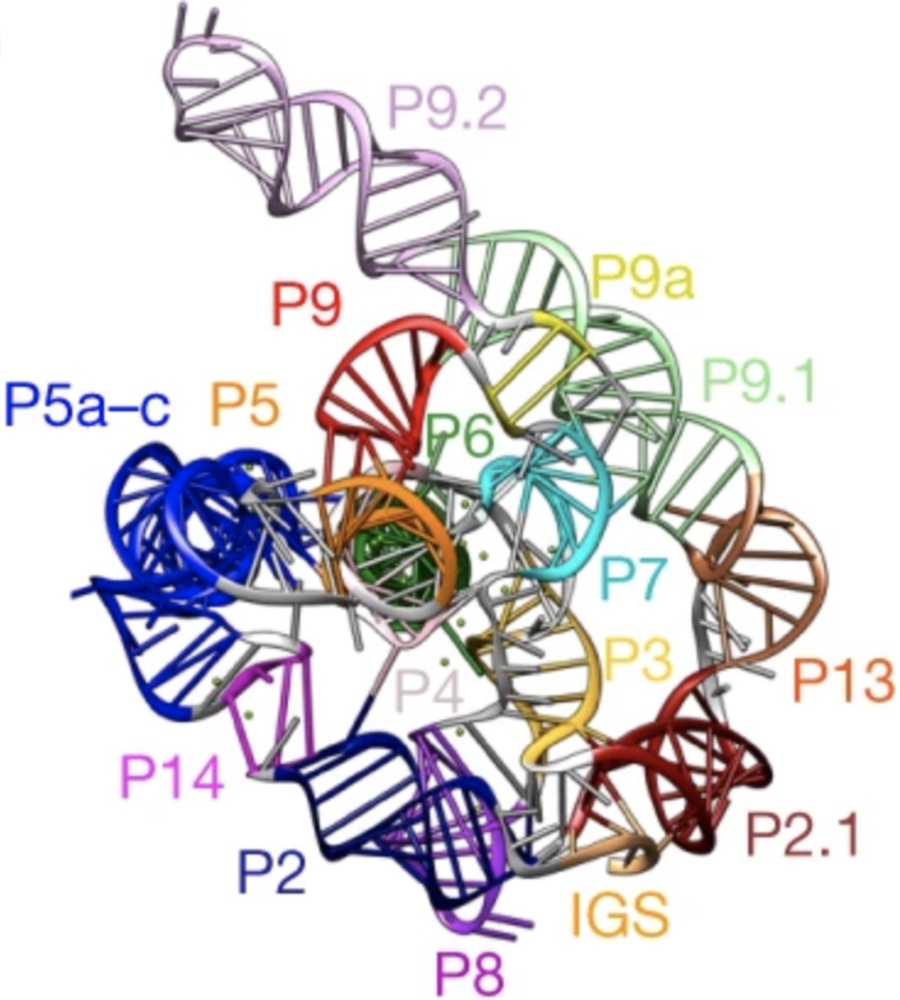
We’ve reported high resolution cryo-EM structures of the full-length Tetrahymena ribozyme in a paper out today. Wonderful to start getting a close look at this paradigmatic RNA-only enzyme, discovered by Tom Cech and colleagues four decades ago! You can read more about our work to rapidly solve RNA structures with Wah Chiu at this SLAC piece.
We’re delighted that Rachael Kretsch will present on RNA cryo-EM at the American Crystallographic Association meeting this week! Rachael’s talk will be on Tuesday, August 3, 2021, as part of the session “2.2.5 What Can & Can’t We See Reliably at Resolution X?”, 12-3pm ET. We are also very happy to celebrate the presentation of the Buerger Award to our wonderful collaborator Wah Chiu. Congratulations!
We were pleased to help host EternaCon 2021, a virtual celebration of Eterna’s efforts in inclusion, ribosome engineering, and mRNA vaccine research, with previews of upcoming developments. Check out what happened, including video of talks, at the EternaCon 2021 page
Congratulations to Hannah for successfully defending her Ph.D. in Chemistry! PhD student Hannah Wayment-Steele has been awarded the Chemical Computing Group excellence award from the American Chemical Society! You can read more about the award here. She will present her work at the ACS national meeting later in 2021. Hannah also spoke recently on work developing computational methods for mRNA stabilization at Schrodinger’s Multiscale Modelling for Biotherapeutics symposium
Congratulations to Rachael Kretsch for being awarded a Bio-X Bowes Stanford Interdisciplinary Graduate Fellowship! The award will support her thesis research on advancing RNA structure determination with cryo-EM and computer modeling, with close collaboration from Wah Chiu.
Congratulations to Ramya Rangan for being awarded a 2021 Gerald J. Lieberman Fellowship! This Stanford-wide fellowship supports PhD students intending to pursue careers in academia who have demonstrated potential for leadership roles through their research accomplishments, teaching and university service.
India is entering into lockdown and there’s hope of developing new mRNA vaccines against emerging regional variants of SARS-CoV-2 like B.1.617. Eterna has launched a design lab to create a superfolder mRNA encoding the “S-2P” spike protein for the B.1.617 variant and raising funds to experimentally test the stability and potency of the top-ranked solutions here at Stanford. Come join us at Eterna.
Our year-long collaboration with Maria Barna’s group and Eterna, which started at the onset of the COVID-19 pandemic, has resulted in a variety of novel methods and principles for designing stabilized mRNA therapeutics. Our first preprint describing our experimental results is up! Check out this Q&A with Maria Barna and Rhiju Das explaining why and how we did this research.
Congratulations to Lexy Strom, senior biology major at San Diego State university and Das lab SSRP student-turned-research-affiliate, who was awarded the NSF Graduate Research Fellowship!
Programmer position available. Worldwide immunization is bottlenecked by the difficulty of distributing mRNA vaccines, whose poor stability currently requires ultra-deep-freeze storage. Through 2020, my lab, Stanford’s OpenVaccine consortium, the Eterna crowdsourcing project, and Kaggle competitors have developed methods to stabilize these mRNA molecules. We are currently limited by the slow speed of RNA secondary structure modeling and design methods, and seek an extremely fast and detail-oriented programmer to rapidly re-code dynamic programming methods by February 2021. This much-needed code will be made available freely under BSD-3 license for all upcoming research on RNA medicines in academia and industry.
Jobs available. We and our collaborators in Maria Barna’s lab are engaged in research to stabilize COVID-19 mRNA vaccines to enable world-wide immunization with collaboration with a leading industry consortium. We are looking for two research assistants with backgrounds in experimental RNA molecular biology to join these efforts ASAP. Please apply at the Stanford jobs website for the Life Sciences Research Professional or Life Sciences Technician position.”
Rhiju spoke on our recent SARS-CoV-2 RNA structural biology research at an upcoming Biophysical Society symposium entitled “Biophysicists Address COVID-19 Challenges” on Thursday, Oct. 29, 2020. See you there! Register for free at the biophysical society website.
Congrats to David Cox, who has been selected to speak at the Stanford.Berkeley.UCSF Next Generation Faculty symposium highlighting exceptional early career scientists! Register for the symposium, which will be on Oct. 23, 2020
Improving the stability of RNA vaccines will be important for worldwide vaccine distribution. We present a biophysical model for predicting RNA degradation in a new preprint led by Hannah Wayment-Steele. Through a variety of mRNA design challenges launched through the OpenVaccine challenge on Eterna by Do Soon Kim, we predict that the stability of any RNA vaccine can be increased at least two-fold either through rational design, as demonstrated by Eterna participants, or algorithmically, through the algorithms RiboTree (developed by Christian Choe) and LinearDesign.
EternaCon 2020 is happening on Saturday-Sunday, July 25-26, 2020! Starts at 9am PDT, 12pm EDT, 4pm UTC. Come (virtually) and hear about the latest Eterna innovations in OpenVaccine and super-folder ribosomes, including talks from Eterna players, world-leading researchers, and Eterna’s latest allies at HBCU Prairie View A&M. Checkout the <href=”https: eternagame.org=”” eternacon=”” 2020″=””>schedule and register to attend virtually.</href=”https:>
We’ve resolved the SARS-CoV-2 frameshift stimulation element with our hybrid structure determination pipeline Ribosolve, and we report effects of antisense oligonucleotides on this RNA’s function and on virus replication in cell culture. Read more in our bioRxiv preprint. Many thanks to our colleagues in Glenn, Chiu, d’Souza, and Baric labs and SLAC for this fruitful collaboration, much of which occurred in the midst of.our university shutdowns.
Which RNA secondary structure package best predicts experimental data? Hannah Wayment-Steele used high-throughput structural data from Eterna to answer this question, and found a suprising result: lesser-known packages developed by statistical learning performed notably better than more widely-used packages. Motivated by these results, she developed a multi-task learning algorithm trained on Eterna data, named EternaFold, which showed the best predictions on Eterna data as well as completely independent datasets of viral RNAs and synthetic mRNAs important for vaccine design. We hope that the EternaFold algorithm will be useful for improving RNA design, and that the EternaBench database will provide an independent platform to benchmark more improvements in secondary structure prediction.
Congratulations to Roger Wellington-Oguri, Eli Fisker, Mathew Zada, Michelle Wiley, Jill Townley, and Eterna Players for the first peer-reviewed scientific publication written entirely by videogame players. The paper “Evidence of an Unusual Poly(A) RNA Signature Detected by High-throughput Chemical Mapping” was just accepted in the journal Biochemistry. The paper is coming out back to back with a companion paper from our lab that validates the players’ discovery of an unusual polyadenosine signature in RNA structure mapping experiments. These same player authors are leading scientific discussions in Eterna’s OpenVaccine mRNA vaccine stabilization efforts
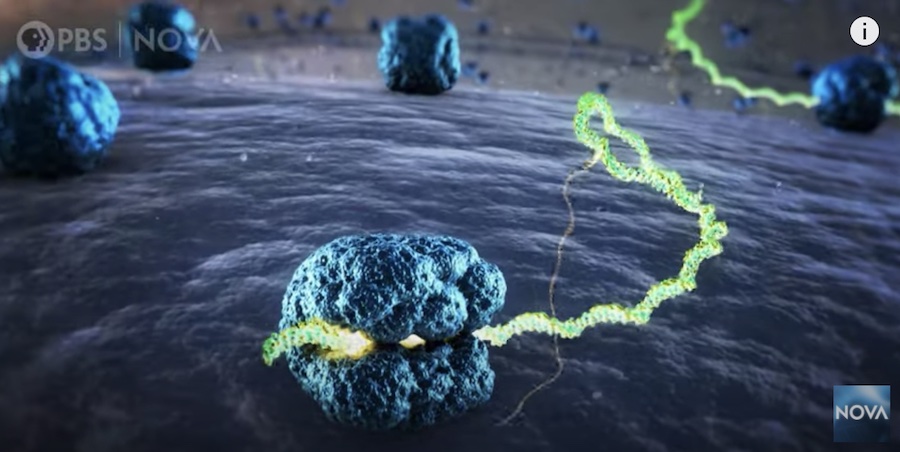
Das lab’s COVID-19 mRNA vaccine stabilization efforts, along with Eterna OpenVaccine, are featured in PBS Nova’s recent Decoding COVID-19 TV special. More about our efforts in the Stanford School of Medicine Scope blog. Rhiju also presented the efforts at the Stanford School of Medicine’s COVID-19 Town Hall. Come play the Eterna game to help us stabilize RNA vaccines at eternagame.org. If you’re interested in a more detailed science seminar given in Stanford’s Frontiers in Biosciences series, please contact Rhiju directly
There are many conserved and likely structured elements of the SARS-CoV-2 genome, such as those identified in a previous preprint, with no known 3D structure, which nonetheless may be useful targets for small molecule therapeutics. Ramya Rangan and Andy Watkins have performed studies with our latest structure prediction code, FARFAR2, to make publicly available collections of de novo 3D models of these elements. Their linked preprint has been made available in bioRxiv.
To aid in the design of nucleic acid-based therapeutics for the raging COVID-19 pandemic, Ramya Rangan and Ivan (Vanya) Zheludev have worked quickly to conduct a comprehensive conservation analysis of the SARS-CoV-2 genome. Their preprint, published today in bioRxiv, identifies in this RNA virus many highly conserved sequences and secondary structure. A follow-up analysis confirms the conservation of many of these regions in the 739 SARS-CoV-2 genomes published during the current outbreak. The regions they have identified provide promising targets for the development of antiviral anti-sense oligonucleotide treatments as we continue to fight the COVID-19 pandemic.
We are proud to launch Eterna’s OpenVaccine challenge. These puzzle seeks improved mRNA sequences for COVID-19 vaccines, with a special focus to stabilizing these molecules against degradation to help enable their deployment for mass immunization. Experimental tests are being carried out in collaboration with Maria Barna’s lab in Stanford Genetics. Come join us!
Our laboratory won the overall prize in the Kornberg memorial Biochemistry department baking contest , for the second year in a row! In addition, Ramya Rangan won first place in the pie category for her blueberry pie, rotation student Jon Doenier took the cake with his chocolate mousse cake, Vanya Zheludhev directed this year’s skit video, and Matt Adrianowycz MC-ed the winning ceremonies. Happy holidays to all and to the entire department!
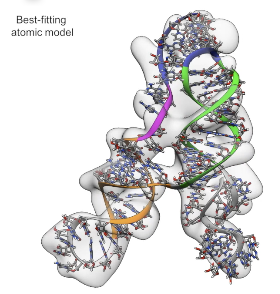
In collaboration with Wah Chiu’s lab, we’re pushing the limits of cryo-EM single-particle analysis to solve structures. An article in Nature Communications describes our results on a 40 kDa RNA, the SAM-IV riboswitch aptamer. The paper includes analyses confirming that the ligand-binding site can be detected in the RNA. Come play the Eterna puzzle celebrating this beautiful molecule (worth 500 points)
After a successful pilot experiments in early 2019, Eterna is releasing Ribosome Challenge Round 1. The goal of this challenge is to create a superfolder variant of the E. coli ribosome whose assembly might be less fragile to mutations or changed solution conditions. If achieved, such a molecule could help worldwide efforts to repurpose the ribosome to make alternative polymers and more. Our initial challenge is making use of algorithmic innovations from Liang Huang (Oregon State University) and rapid in vitro assembly measurements by the lab of Mike Jewett (Northwestern University), as well as new hand-designed layouts of the ribosome RNA in two dimensions that match its three-dimensional structure. This lab round collects solutions on Feb. 1, 2020 — and remember that you need to play the incoming puzzle progression to enter the lab and take part in the discussion and solving.
Congratulations to Kalli Kappel for winning the American Chemical Society’s 2019 Chemical Computing Group excellence award for graduate students. She will be presenting at the ACS meeting in San Diego in late August, on a hybrid method for rapid experimental RNA structure determination called Ribosolve
The fifth annual EternaCon was held at Stanford University on July 20-21 2019! Much of our lab was there as we discuss the next ambitious moonshots for this exciting project. Check out the schedule of events at http://eternawiki.org/wiki/index.php5/Eternacon.
Rhiju spoke about “Computational Design of RNA Structure/Function” at the Gordon Research Conference on Computer-aided Drug Design, describing our latest studies and inventions on new design principles for RNA therapeutics.
Rhiju spoke at the 2019 Synthetic Biology: Engineering, Evolution & Design (SEED) in the “Cell Free Synthetic Biology” session on June 23-27, 2019. The topic: a new kind of molecular calculator developed to enable inexpensive diagnostics for active tuberculosis, sepsis, and other diseases tied to ratiometric gene signatures.
Rhiju spoke at the RNA society conference in Krakow, Poland on June 11-16, 2019. He presented a new Ribosolve method to solve RNA 3D structures rapidly.
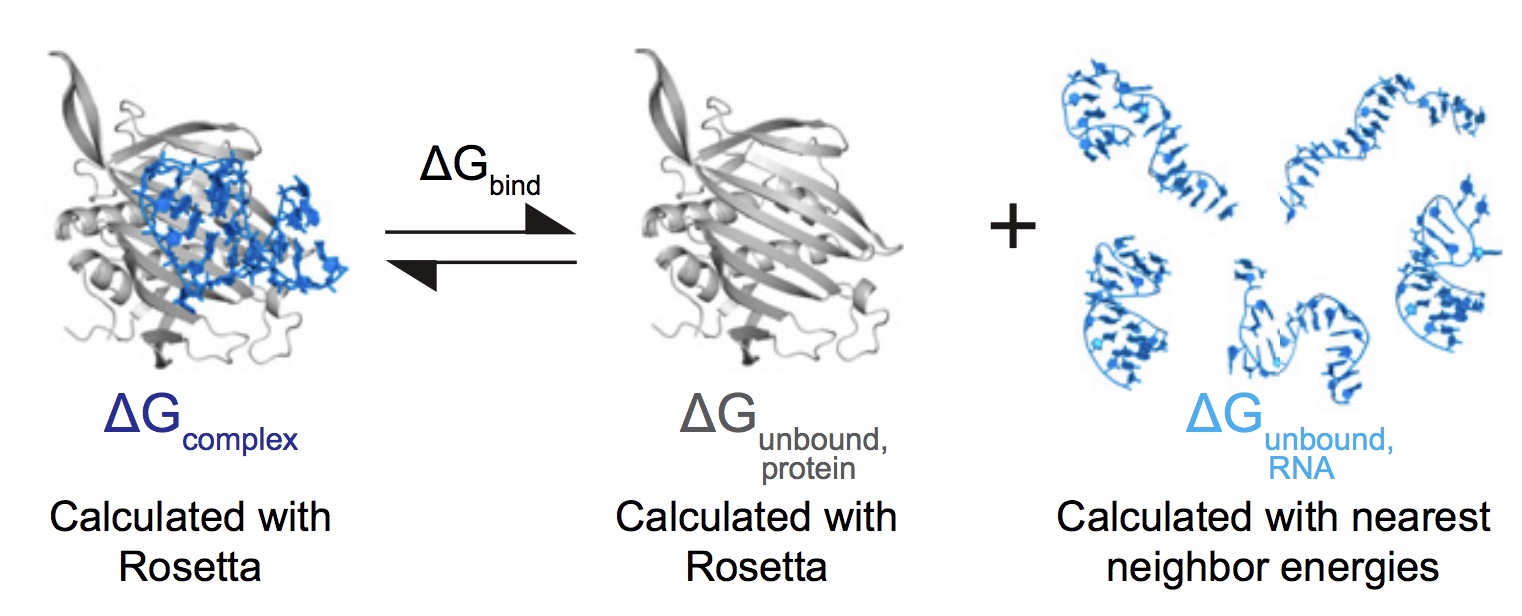
The latest paper by Kalli Kappel, on a Rosetta computational infrastructure for modeling energetics of RNA-protein complexes, has been published in PNAS. Check out Kalli’s other recent papers on RNA-protein structure modeling with Rosetta in Structure and Nature Methods!
Ann Kladwang celebrates her 10th anniversary as a research associate in the Das laboratory. Congratulations and thanks, Ann, from everyone who has learned RNA experiments from you!
Matt Adrianowycz was awarded a Rosetta Commons grant for ‘Rapid free energy calculations in Rosetta’! Congratulations Matt!
Kalli Kappel gave a talk titled ‘Structure determination of RNA and RNA-protein complexes through cryo-EM and computational modeling’ at the Bay Area RNA Club’s yearly meeting at the University of California, San Francisco
Congratulations to Das laboratory for winning two categories in the annual Biochemistry department holiday baking championship as well as the grand prize! Not bad for a bunch of mostly theorists. Dr. Michael Gotrik won for an ollallieberry cheesecake, and Dr. Feriel Melaine won for an apple pie with a French twist
The fourth annual EternaCon is happening on Saturday, August 11, and Sunday, August 12, 2018 at Stanford University. Come join us at Li Ka Shing Center (Room 120) or tune in online. As in prior years, the event is fully organized by Eterna players, and most speakers and panels are by players. We also welcome Liang Huang, Dave Hendrix, and Ingmar Riedel-Kruse as scientist guest speakers. More information and late registrations available through Eterna, and a draft schedule is available here
Kalli gave an oral presentation entitled De novo computational RNA modeling into cryoEM maps of large ribonucleoprotein complexes at the 23rd annual RNA Society meeting at the University of California
Congratulations to Matt Adrianowycz for winning a Stanford Centennial Teaching Award! Remarkably, Matt has won the award as a first-year Ph.D. student, for his teaching in Chemistry 141: The Chemical Principles of Life.
Rhiju spoke at Tsinghua University at the 2018 Tsinghua International Symposium of Computational Structural Biology and Biophysics, on Eterna’s recent progress in RNA design of on-demand molecular calculators. More information about the event can be found here.
Kate Coppess has won a 2018 NSF graduate research fellowship award! Kate is also a Stanford EDGE-STEM fellow and co-president of Graduate Students of Applied Physics and Physics Congratulations, Kate!
Our lab and Eterna are piloting a new form of peer review for scientific publications. Because so many of our upcoming papers will involve players as co-authors, we wanted to find a way for anyone to leave & respond to comments on papers before and during paper review, in the nearly frictionless way that Google docs allow. So that’s what we’re doing. We are making papers available on preprint servers like arXiv and bioRxiv and we are including a Google doc link in the abstract that players (or other members of the public) can follow. We can all leave comments — and comments on comments — in that Google doc. These comments will show up as notifications to the paper’s lead author, who can join those threads and quickly make edits to the paper. As a pilot, Rhiju is putting up a paper on a data analysis method for quantifying thermodynamics in RNA structure mapping experiments. Check it out on the LIFFT BioRxiv preprint and the LIFFT community review Google doc . An Eterna blog post is here
The Das & Greenleaf labs at Stanford University School of Medicine are seeking ambitious postdoctoral researchers to create the next generation of the Eterna internet-scale videogame for RNA design. Projects involve ribosome redesign, RNA-based diagnostics, controllable genome editing, and the origin of life. More info on position and how to apply at Nature Jobs
Kalli Kappel spoke at the 4th International Conference on Protein and RNA Structure Prediction! She presented her latest work on Computational modeling of missing RNA coordinates into density maps of ribonucleoprotein machines.
We’re pleased to report our latest methods in 3D RNA modeling and Eterna design. Three preprints on BioRxiv highlight Blind prediction of noncanonical RNA structure at atomic accuracy, Computational design of asymmetric three-dimensional RNA structures and machines, and Prospects for recurrent neural network models to learn RNA biophysics from high-throughput data. Check out preprints through our publications page
Congratulations to collaborators Tosan Omabegho & Zev Bryant for reporting their design of a fast molecular machine whose direction can be reversed back and forth by RNA. The paper “Controllable molecular motors engineered from myosin and RNA” and a commentary are now out in Nature Nanotechnology
A rousing EternaCon happened on 2017 Saturday and Sunday October 21-22, 2017. Nearly 50 Eterna players and experts in CRISPR technologies and inexpensive diagnostics came together to consider the possibility of citizen scientists inventing medicine.
Eterna has launched the OpenCRISPR challenge, in partnership with Stanford’s Center for Personal Dynamic Regulomes and Berkeley’s Innovative Genome Institute. These Eterna puzzles involve designing on/off switches for CRISPR that will be tested experimentally. We’re hoping for 100,000 solutions! Follow us on Twitter @eternagame
The Das lab at Stanford is hiring. One position is for an information analytics aide who can work approximately 20 hours/week and who lives sufficiently close to the Bay Area to come into Stanford one day of the week. The work is well suited to undergraduates who are pursuing computational biology degrees. Eterna players are particularly encouraged to apply.
Our latest papers take us closer to multidimensional chemical mapping of RNA in vivo and in complex structural ensembles. You can read about these methods before publication through preprints on bioRxiv: RNA structure inference through chemical mapping after accidental or intentional mutations and Allosteric logic of the V. vulnificus adenine riboswitch resolved by four-dimensional chemical mapping. Update: the M2-seq paper is out in PNAS
Michelle Wu spoke at the ISMB RNA-SIG meeting in Prague, Czech Republic. She presented work from our lab and Eterna on “Recurrent neural network models of RNA interactions”
Johan Andreasson presented a talk from the Eterna project reporting the discovery of “Thermodynamically optimal riboswitches through massively parallel design” at the RNA 2017 conference in Prague, Czech republic. Collaborators Marcin Magnus (Bujnicki lab) and Brant Gracia (Russell lab) will also be giving talks on work involving computational and experimental methods from the Das lab.
Joe Yesselman reported his work on “Automated Design of Three-Dimensional Asymmetric RNA Structures at Near-Atomic Accuracy” in an invited talk at FNANO 2017 (Foundation of Nanoscience)!
Rhiju presented recent progress in ‘RNA puzzles’ at a Cell Press webinar “Predicting and Observing RNA Folding: A Progress Report” along with Kathleen Hall, Sarah Woodon, and Rob Batey. The web cast is available at the Cell website.
Congratulations to Kalli Kappel for winning a travel award to the 2017 Biophysical Society annual meeting, in New Orleans, LA. She presented her work on “Blind predictions of RNA/protein relative binding affinities”, and won a best poster prize!
Congratulations to Andy Watkins for receiving a Dean’s Fellowship to support his early postdoctoral work in the Das lab! Andy joins us after completing his Ph.D. from the Arora and Bonneau labs at NYU.
Siqi Tian successfully defended his Ph.D. thesis on “Robust Validation of RNA Secondary Structure & Ensembles through Multidimensional Chemical Mapping” on September 19, 2016.
Congratulations to Clarence Yu Cheng for successfully defending his Ph.D. thesis on “Advancing multidimensional chemical mapping of RNA structure through next-generation sequencing”.
Get ready for Eternacon 2016! This year’s meeting will take place at Stanford University on Saturday and Sunday, July 16-17. For more information, please visit the Eternacon 2016 wiki.
Congratulations to Kalli Kappel for receiving a Grace Hopper Celebration Scholarship! She will represent our lab at the GHC on October 16, 2016.
Eterna is featured in Werner Herzog’s new film “Lo And Behold: Reveries of the Connected World”. Check out the new trailer!
We are launching a beta version of a new site describing workflows through all the RNA software and experimental resources that we’ve developed. Do you want to learn an RNA’s secondary structure? Model its tertiary structure? Come check out RiboKit and send us feedback.
Upcoming: Dr. Joe Yesselman will represent our lab at the international RNA 2016 conference in Kyoto, Japan, which starts on July 28, 2016. He will present collaborative work with the Greenleaf and Herschlag labs showing that “Surprisingly Accurate Energetic Modeling Deconvolves Big Data on RNA 3D Folding”.
Stanford magazine writes about our lab and recent developments in Eterna, including the player-created Eterna100 benchmark and the OpenTB challenge.
Eterna has launched the OpenTB challenge, a collaboration with Purvesh Khatri’s bioinformatics lab and Will Greenleaf’s genetics lab to develop new molecules for tuberculosis diagnostics. Come play eterna to take part in the game. Press accounts are at Wall Street Journal, including multimedia, and NPR
Eterna citizen scientists are lead authors on peer-reviewed paper “Principles for Predicting RNA Secondary Structure Design Difficulty“. Press coverage includes Science Magazine, Washington Post, Reuters.
elNando888 and Ben Keep presented Eterna at the AAAS 2016 annual meeting, in the session on “Massively-Collaborative Global Research in Mathematics and Science”.
Clarence Cheng presented an Eterna open group meeting on “MaP-2D: Extracting two-dimensional structural information from mutational profiling experiments”, on February 15, 2016. Tune into our eterna YouTube channel for this and previous talks.
Joseph Yesselman presented an EteRNA open group meeting on RNAmake and the future of eterna 3d
Kalli Kappel presented an EteRNA open group meeting on Predictions of RNA/protein binding affinities: Optimization and validation of a general score function for RNA/protein interactions
Michelle Wu presented an EteRNA open group meeting on upcoming Eterna medicine initiatives, on November 6, 2015. Tune into our eterna YouTube channel for this and previous talks.
Vineet Kosaraju was awarded the 2015 Davidson fellowship for his “3D RNA Engineering in a Massive Open Laboratory” project on Eterna. Congratulations, Vineet!
Johan Andreasson presented an EteRNA open group meeting on new result from riboswitch labs, on October 9, 2015. Tune into our eterna YouTube channel for this and previous talks.
RNA lab (a joint collaboration of Eterna and NOVA) is a marquee banner app in the Chrome Web store. And RNA lab is now part of NOVA’s partner collection in Khan Academy.
The first ever EternaCon happened at Stanford University on July 11-12! Video and proceedings are being collected at the Eterna site.
Pablo Cordero gave his final thesis talk on “Using factorization methods to find structure in RNA molecules and networks”. Congratulations, Pablo!
Fang-Chieh Chou defended his thesis on “Advancing high resolution predictions of RNA structure and energetics”. Congratulations, Fang!
Joe Yesselman presented an EteRNA open group meeting on new tools for RNA 3D design, on June 5, 2015. Tune into our eterna YouTube channel for this and previous talks
Rhiju to receive 2015 OpenEye Outstanding Junior Faculty Award in Computational Chemistry!
Rhiju will give a keynote at DNA20, the 20th international conference on DNA computing and molecular programming, in Kyoto, Japan.
Rhiju & Fang recognized with the CLASHBUSTERS! award from the lab of Jane & Dave Richardson at the RNA/protein session of RosettaCon 2014. And Joe Yesselman won best poster of the conference for “Towards Automated Design of 3D RNA Structure”!
Clarence Cheng presented a new technology for solving RNA 3D folds through multiplexed hydroxyl radical cleavage analysis (MOHCA-seq) at the international RNA society meeting on June 3-8, 2014 in Quebec City, Canada. Congratulations to Clarence for being selected to give a talk in the High Throughput methods session. Rhiju also presented on EteRNA at the Bioinformatics Workshop at the meeting.
Newly discovered mushroom named after lab member! Congratulations to Wipapat (Ann) Kladwang for co-discovering Pseudobaeospora wipapatiae, described in “A ruby-colored Pseudobaeospora species is described as new from material collected on the island of Hawaii”, in the newest issue of Mycologia.
Rhiju Das presented on RNA Structure: Think Big” on May 9 at 9am PST. Join us interactively and be sure to stay tuned to news on EteRNA for more information. We will monitor EteRNA chat for real-time questions. Missed our prior monthly open group meetings? Check them out on our YouTube channel for one month after broadcast.”
Congratulations to the PBS NovaLab/EteRNA team for creating NOVA’s RNA VirtuaLab, interactive content on RNA and molecular biology for US schools.
First scientific paper from EteRNA, “RNA Design Rules from a Massive Open Laboratory”, published in PNAS. A nice highlight in Science magazine was also published here: Online Video Game Plugs Players Into Remote-Controlled Biochemistry Lab.
Das Lab initiates our monthly ‘open group meeting’, focused on science happening on EteRNA. See our first meeting here, and join us interactively at our next meetings, happening monthly. Stay tuned to news on EteRNA for more information.
Presenting our new fixed backbone design server for RNA. Want to know what sequence best fits a given phosphate backbone? Come try our server and submit any RNA in PDB format.
Parin Sripakdeevong, Ph.D. student in biophysics, will be defending his thesis on Modeling noncanonical RNA 3D structures at atomic resolution under the Rosetta framework on Monday, Nov 18th, 2013: 3:00 PM – 4:00 PM in Clark S360.
A NOVA feature on our colleague Adrien Treuille and the EteRNA project has won a Kavli award for scientific reporting. Congratulations to Adrien and to director Joshua Seftel!
Rhiju gave a keynote at the Design Research Conference in Chicago on Oct. 8, 2013. The 12th annual Design Research Conference hosted by IIT Institute of Design will explore design’s role in mediating these forces: Are they opposable? Synthesized? Unified? Practicing designers, artists, scientists, engineers, educators, and entrepreneurs will present a variety of perspectives on balancing multiple creative forces amidst complex systems and issues.



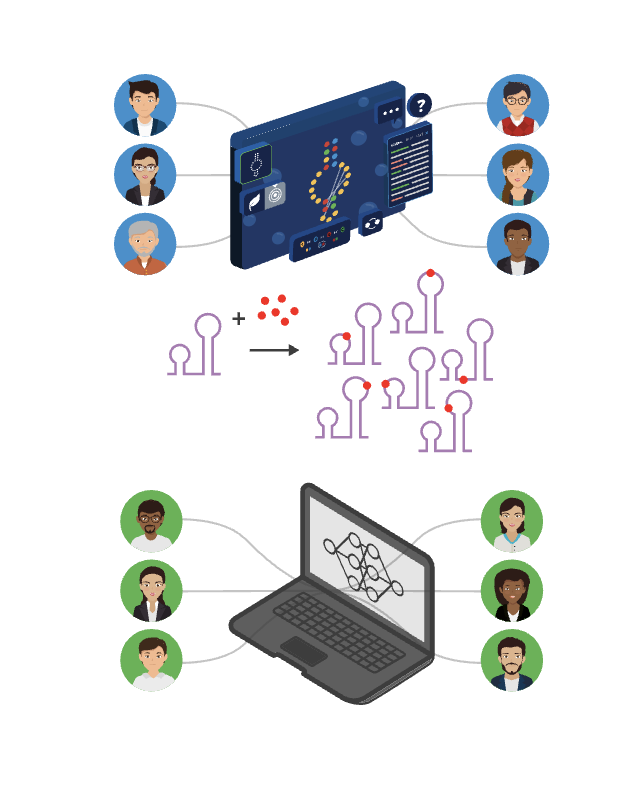

![Call to action to RNA and ML folks everywhere: We are getting ready to scale up our chemical mapping data protocol, initially to 8M sequences, and then onwards to 100 M and, hopefully 1B, sequences. For this Ribonanza 2 initiative, we’re looking for diverse sets of RNA molecules from as many different groups as possible! Do you have a list of natural or designed RNA sequences that you’re curious to visualize? We collected the first round in January 15, 2024, and if you’re interested in joining the next round, e-mail us at ribonanza [at] stanford.edu.](https://das-lab.staging1090.pendari.com/wp-content/uploads/2024/08/thumb76_76-1.png)