arrowSearch for papers from the Das Lab on PubMed
2024
 He, S., Huang, R., Townley, J., C. Kretsch, R., Karagianes, T.G., Cox, D.B.T., Blair, H., Penzar, D., Vyaltsev, V., Aristova, E., Zinkevich, A., Bakulin, A., Sohn, H., Krstevski, D., Fukui, T., Tatematsu, F., Uchida, Y., Jang, D., Seong Lee, J., Shieh, R., Ma, T., Martynov, E., Shugaev, M.V., Bukhari, H.S.T., Fujikawa, K., Onodera, K., Henkel, C., Ron, S., Romano, J., Nicol, J.J. , Nye, G.P., Wu, Y., Choe, C., Reade, W., Eterna participants., Das, R. (2024)
He, S., Huang, R., Townley, J., C. Kretsch, R., Karagianes, T.G., Cox, D.B.T., Blair, H., Penzar, D., Vyaltsev, V., Aristova, E., Zinkevich, A., Bakulin, A., Sohn, H., Krstevski, D., Fukui, T., Tatematsu, F., Uchida, Y., Jang, D., Seong Lee, J., Shieh, R., Ma, T., Martynov, E., Shugaev, M.V., Bukhari, H.S.T., Fujikawa, K., Onodera, K., Henkel, C., Ron, S., Romano, J., Nicol, J.J. , Nye, G.P., Wu, Y., Choe, C., Reade, W., Eterna participants., Das, R. (2024)
“Ribonanza: deep learning of RNA structure through dual crowdsourcing”
bioRxiv
Preprint | Link
Choe, C.A., Andreasson, J.O.L., Melaine, F., Kladwang, W., Wu, M.J., Portela, F., Wellington-Oguri, R., Nicol, J.J., Wayment-Steele, H.K., Gotrik, M., Eterna Participants, Khatri, P., Greenleaf, W.J., Das, R. (2024) “Compact RNA sensors for increasingly complex functions of multiple inputs” bioRxiv | Paper | Preprint
2023
Kretsch, R., Andersen, E., Bujnicki, J., Chiu, W., Das, R., Luo, B., Masquida, B., McRae, E., Schroeder, G., Su, Z., Wedekind, J., Xu, L., Zhang, K., Zheludev, I., Moult, J., Kryshtafovych, A. (2023) “RNA target highlights in CASP15: Evaluation of predicted models by structure providers” Proteins: Structure, Function, and Bioinformatics 91 (12) : 1600 – 1615 | Paper | Link
Kryshtafovych, A., Antczak, M., Szachniuk, M., Zok, T., Kretsch, R., Rangan, R., Pham, P., Das, R., Robin, X., Studer, G., Durairaj, J., Eberhardt, J., Sweeney, A., Topf, M., Schwede, T., Fidelis, K., Moult, J. (2023) “New prediction categories in CASP15” Proteins: Structure, Function, and Bioinformatics 91 (12) : 1550 – 1557 | Paper | Link
Krüger, A, Watkins A.M., Wellington-Oguri R., Romano, J., Kofman, C., DeFoe, A., Kim, Y., Anderson-Lee, J., Fisker, E., Townley J., Eterna Participants, d’Aquino, A.E., Das, R., Jewett, M.C. (2023) “Community science designed ribosomes with beneficial phenotypes” Nature Communications 14 : 961 | Paper | Link
2022
 Wayment-Steele H., Kladwang W., Watkins A., Kim D., Tunguz B., Reade W., Demkin M., Romano J., Wellington-Oguri R., Nicol J., Gao J., Onodera K., Fujikawa K., Mao H., Vandewiele G., Tinti M., Steenwinckel B., Ito T., Noumi T., He S., Ishi K., Lee Y., Öztürk F., Chiu K., Öztürk E., Amer K., Fares M., Eterna Participants , Das R. (2022)
Wayment-Steele H., Kladwang W., Watkins A., Kim D., Tunguz B., Reade W., Demkin M., Romano J., Wellington-Oguri R., Nicol J., Gao J., Onodera K., Fujikawa K., Mao H., Vandewiele G., Tinti M., Steenwinckel B., Ito T., Noumi T., He S., Ishi K., Lee Y., Öztürk F., Chiu K., Öztürk E., Amer K., Fares M., Eterna Participants , Das R. (2022)
“Deep learning models for predicting RNA degradation via dual crowdsourcing”
Nature Machine Intelligence 4, 1174–1184
Paper | Link
 Wayment-Steele, H.K., Kladwang, W., Strom, A.I., Lee, J., Treuille, A., Becka, A., Eterna Participants, Das R. (2022)
Wayment-Steele, H.K., Kladwang, W., Strom, A.I., Lee, J., Treuille, A., Becka, A., Eterna Participants, Das R. (2022)
“RNA secondary structure packages ranked and improved by high-throughput experiments”
Nature Methods 19 : 1234 – 1242
Paper | Link | EternaBench repository | EternaFold server
Hagey, R.J., Elazar, M., Pham, E.A., Tian, S., Ben-Avi, L., Bernardin-Souibgui, C., Yee, M.F., Moreira, F.R., Rabinovitch, M.V., Meganck, R.M., Fram, B., Beck, A., Gibson, S.A., Lam, G., Devera, J., Kladwang, W., Nguyen, K., Xiong, A., Schaffert, S., Avisar, T., Liu, P., Rustagi, A., Fichtenbaum, C.J., Pang, P.S., Khatri, P., Tseng, C., Taubenberger, J.K., Blish, C.A., Hurst, B.L., Sheahan, T.P., Das, R., Glenn, J.S. (2022) “Programmable antivirals targeting critical conserved viral RNA secondary structures from influenza A virus and SARS-CoV-2” Nature Medicine 28 (9) : 1944 – 1955 | Paper | Link
 Leppek, K.*, Byeon, G.W.*, Kladwang, W.*, Wayment-Steele, H.K.*, Kerr, C.H.*, Xu, A.F.*, Kim, D.S.*, Topkar, V.V., Choe, C., Rothschild, D., Tiu, G.C., Wellington-Oguri, R., Fujii, K., Sharma, E., Watkins, A.M., Nicol, J.J., Romano, J., Tunguz, B., Diaz F., Cai, H., Guo, P., Wu, J., Meng, F., Shi, S., Eterna Participants, Dormitzer, P.R., Solórzano, A., Barna, M., Das, R. *These authors contributed equally. (2022)
Leppek, K.*, Byeon, G.W.*, Kladwang, W.*, Wayment-Steele, H.K.*, Kerr, C.H.*, Xu, A.F.*, Kim, D.S.*, Topkar, V.V., Choe, C., Rothschild, D., Tiu, G.C., Wellington-Oguri, R., Fujii, K., Sharma, E., Watkins, A.M., Nicol, J.J., Romano, J., Tunguz, B., Diaz F., Cai, H., Guo, P., Wu, J., Meng, F., Shi, S., Eterna Participants, Dormitzer, P.R., Solórzano, A., Barna, M., Das, R. *These authors contributed equally. (2022)
“Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics”
Nature Communications 13 : 1536
Paper | Link
2021
 Wayment-Steele, H.K., Kim, D.S., Choe, C.A., Nicol, J.J., Wellington-Oguri, R., Sperberg, R.A.P., Huang, P., Eterna Participants, Das, R. (2021)
Wayment-Steele, H.K., Kim, D.S., Choe, C.A., Nicol, J.J., Wellington-Oguri, R., Sperberg, R.A.P., Huang, P., Eterna Participants, Das, R. (2021)
“Theoretical basis for stabilizing messenger RNA through secondary structure design”
Nucleic Acids Research 49 (18) : 10604 – 10617
Paper | Link | Data (OpenVaccine-solves)
Wayment-Steele, H.K., Kladwang, W., Watkins, A.M., Kim, D.S., Tunguz, B., Reade, W., Demkin, M., Romano, J., Wellington-Oguri, R., Nicol, J.J., Jiayang, G., Kazuki, O., Kujikawa, K., Mao, H., Vandewiele, G., Tinti, M., Steenwinckel, B., Ito, T., Noumi, T., He, S., Ishi, K., Lee, Y., Ozturk, F., Chiu, A., Ozturk, E., Amer, K., Fares, M., Eterna Participants, Das, R. (2021) “Predictive models of RNA degradation through dual crowdsourcing” arXiv
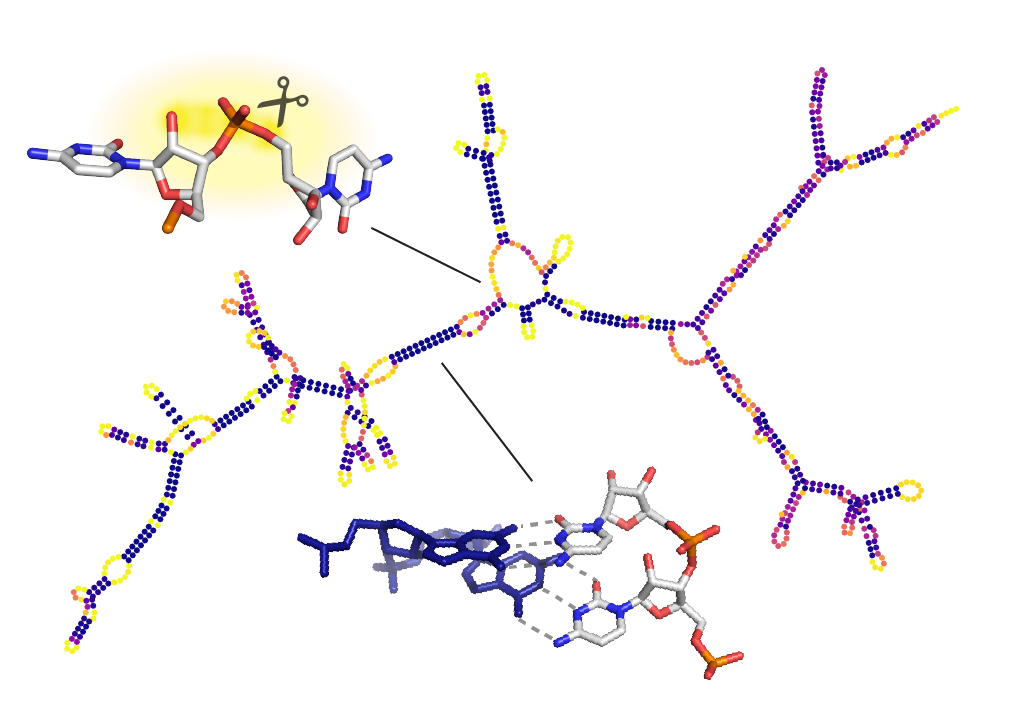
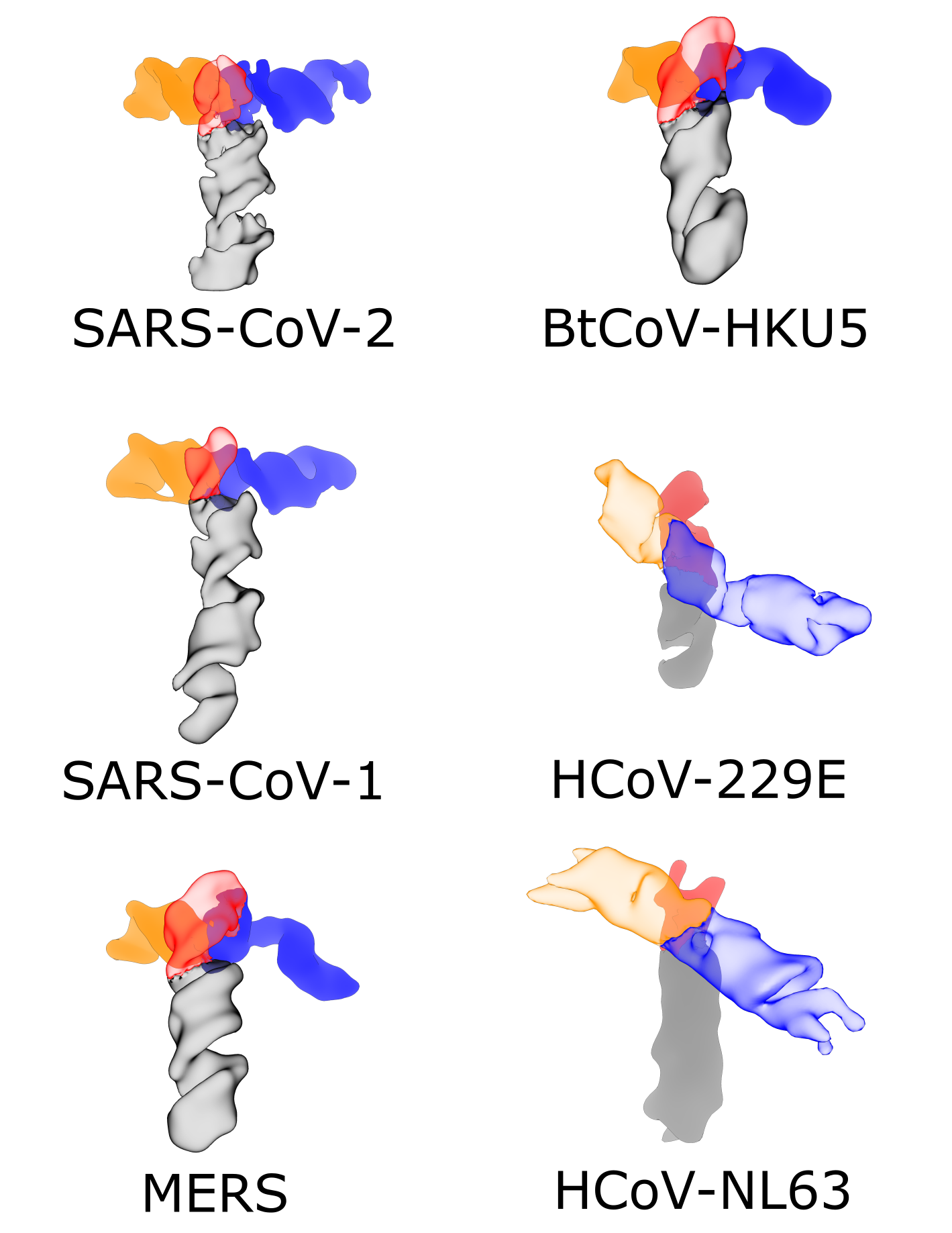
 Zhang, K., Zheludev, I. N., Hagey, R. J., Wu, M. T.-P., Haslecker, R., Hou, Y. J., Kretsch, R., Pintilie, G. D., Rangan, R., Kladwang, W., Li, S., Pham, E. A., Bernardin-Souibgui, C., Baric, R. S., Sheahan, T. P., D Souza, V., Glenn, J. S., Chiu, W., Das, R. (2021)
Zhang, K., Zheludev, I. N., Hagey, R. J., Wu, M. T.-P., Haslecker, R., Hou, Y. J., Kretsch, R., Pintilie, G. D., Rangan, R., Kladwang, W., Li, S., Pham, E. A., Bernardin-Souibgui, C., Baric, R. S., Sheahan, T. P., D Souza, V., Glenn, J. S., Chiu, W., Das, R. (2021)
“Cryo-EM and antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome”
Nature Structure and Molecular Biology 28 : 747 – 754
Paper | Link
Le, K., Adolf-Bryfogle, J., Klima, J., Lyskov, S., Labonte, J., Bertolani, S., Roy Burman, S., Leaver-Fay, A., Weitzner, B., Maguire, J., Rangan, R., Adrianowycz, M., Alford, R., Adal, A., Nance, M., Das, R., Dunbrack, R., Schief, W., Kuhlman, B., Siegel, J., Gray, J. (2021) “PyRosetta jupyter notebooks teach biomolecular structure prediction and design” The Biophysicist 2 (1) : 108 – 122 | Paper | Link
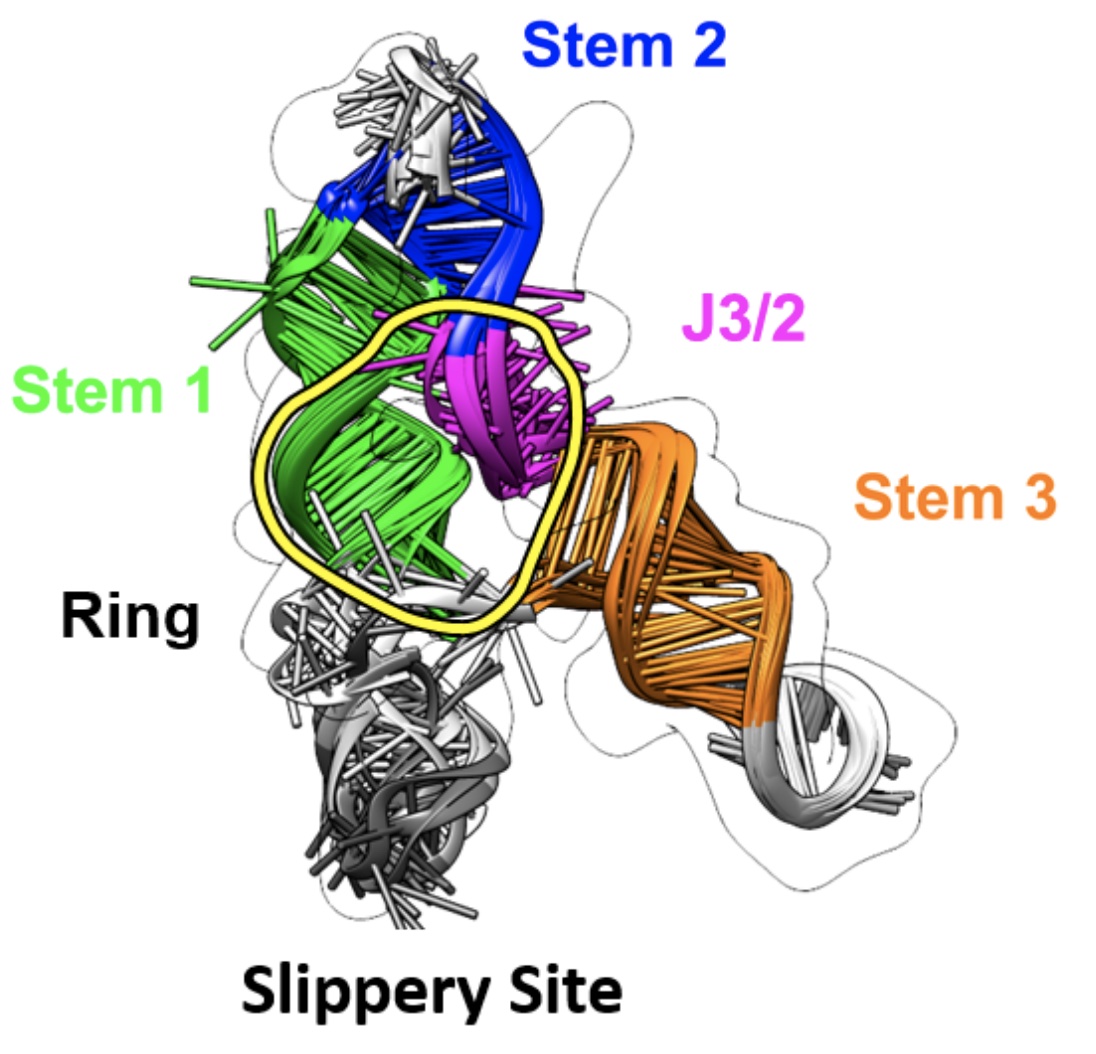
Rangan, R., Watkins, A. M., Chacon, J., Kretsch, R., Kladwang, W., Zheludev, I.N., Townley, J., Rynge, M., Thain, G., Das, R. (2021) “De novo 3D models of SARS-CoV-2 RNA elements from consensus experimental secondary structures” Nucleic Acids Research 49 (6) : 3092 – 3108 | Paper | Link | Data (Model collection, SARS-CoV-2 structures) | Data (Model collection, positive control structures)
2020
Leman, J.K., Weitzner, B.D., Lewis, S.M., Adolf-Bryfogle, J., Alam, N., Alford, R.F., Aprahamian, M., Baker, D., Barlow, K.A., Barth, P., Basanta, B., Bender, B.J., Blacklock, K., Bonet, J., Boyken, S.E., Bradley, P., Bystroff, C., Conway, P., Cooper, S., Correia, B.E., Coventry, B., Das, R., De Jong, R.M., DiMaio, F., Dsilva, L., Dunbrack, R., Ford, A.S., Frenz, B., Fu, D.Y., Geniesse, C., Goldschmidt, L., Gowthaman, R., Gray, J.J., Gront, D., Guffy, S., Horowitz, S., Huang, P.S., Huber, T., Jacobs, T.M., Jeliazkov, J.R., Johnson, D.K., Kappel, K., Karanicolas, J., Khakzad, H., Khar, K.R., Khare, S.D., Khatib, F., Khramushin, A., King, I.C., Kleffner, R., Koepnick, B., Kortemme, T., Kuenze, G., Kuhlman, B., Kuroda, D., Labonte, J.W., Lai, J.K., Lapidoth, G., Leaver-Fay, A., Lindert, S., Linsky, T., London, N., Lubin, J.H., Lyskov, S., Maguire, J., Malmström, L., Marcos, E., Marcu, O., Marze, N.A., Meiler, J., Moretti, R., Mulligan, V.K., Nerli, S., Norn, C., Ó’Conchúir, S., Ollikainen, N., Ovchinnikov, S., Pacella, M.S., Pan, X., Park, H., Pavlovicz, R.E., Pethe, M., Pierce, B.G., Pilla, K.B., Raveh, B., Renfrew, P.D., Burman, S.S.R., Rubenstein, A., Sauer, M.F., Scheck, A., Schief, W., Schueler-Furman, O., Sedan, Y., Sevy, A.M., Sgourakis, N.G., Shi, L., Siegel, J.B., Silva, D.A., Smith, S., Song, Y., Stein, A., Szegedy, M., Teets, F.D., Thyme, S.B., Wang, R.Y., Watkins, A., Zimmerman, L., Bonneau, R. (2020) “Macromolecular modeling and design in Rosetta: recent methods and frameworks.” Nature Methods 17 (7) : 665 – 680
Watkins, A.M., Rangan, R., Das, R. (2020) “FARFAR2: Improved de novo Rosetta prediction of complex global RNA folds” Structure 28 (8) : 963 – 976 | Paper | Link | Server | Benchmarking scripts
Miao, Z., Adamiak, R. W., Antczak, M., Boniecki, M. J., Bujnicki, J., Chen, S.-J., Cheng, C. Y., Cheng, Y., Chou, F.-C., Das, R., Dokholyan, N. V., Ding, F., Geniesse, C., Jiang, Y., Joshi, A., Krokhotin, A., Magnus, M., Mailhot, O., Major, F., Mann, T. H., Piatkowski, P., Pluta, R., Popenda, M., Sarzynska, J., Sun, L., Szachniuk, M., Tian, S., Wang, J., Wang, J., Watkins, A. M., Wiedemann, J., Xiao, Y., Xu, X., Yesselman, J. D., Zhang, D., Zhang, Y., Zhang, Z., Zhao, C., Zhao, P., Zhou, Y., Zok, T., Zyla, A., Ren, A., Batey, R. T., Golden, B. L., Huang, L., Lilley, D. M. J., Liu, Y., Patel, D. J., Westhof, E. (2020) “RNA-Puzzles Round IV: 3D structure predictions of four ribozymes and two aptamers” RNA 26 : 982 – 995 | Paper | Link
2019
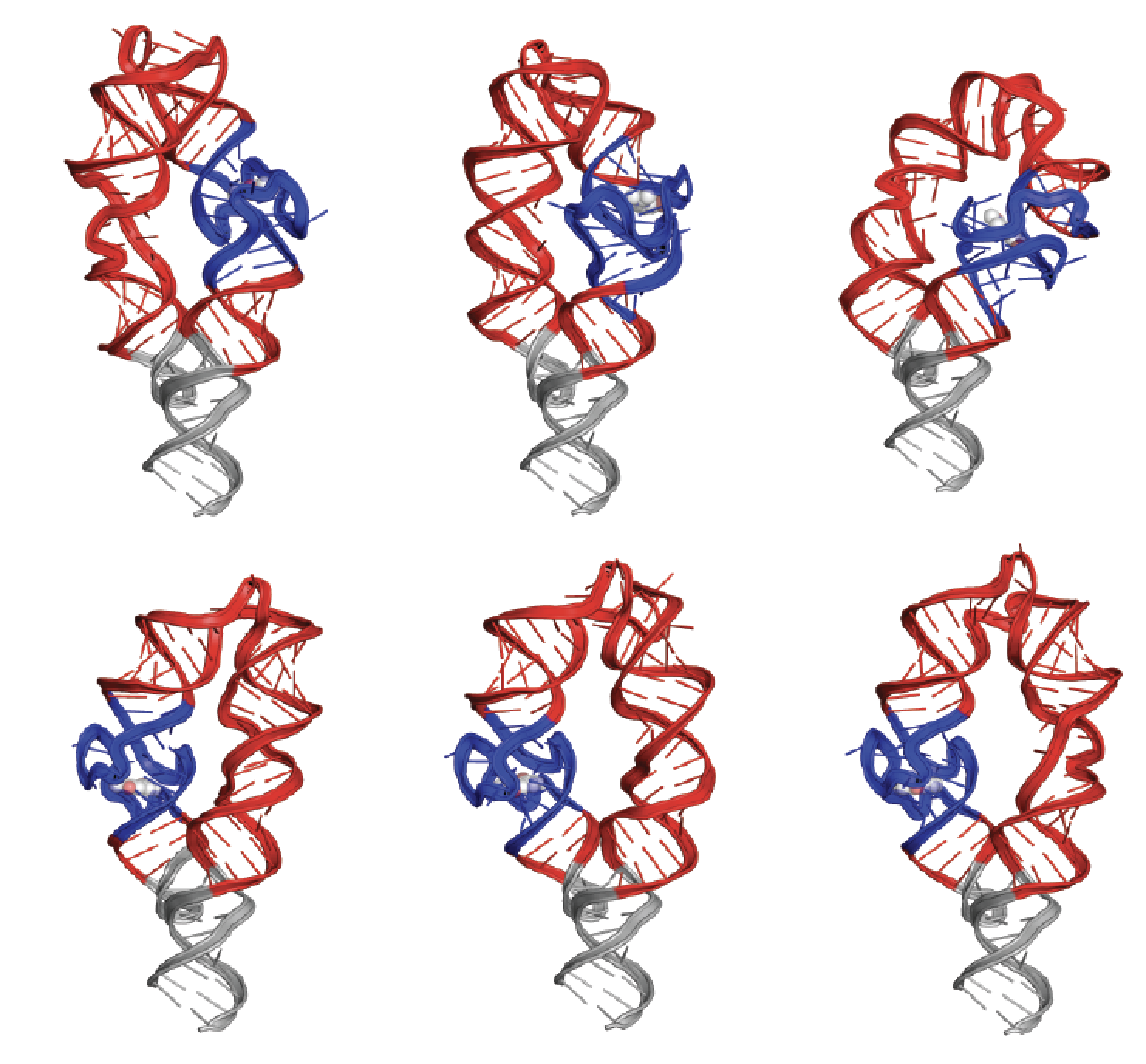 Yesselman, J.D., Eiler, D., Carlson, E.D., Gotrik, M.R., d’Aquino, A.E., Ooms, A.N., Kladwang, W., Carlson, P.D., Shi, X., Costantino, D., Herschlag, D., Lucks, J.B., Jewett, M.C., Kieft, J.S., and Das, R. (2019)
Yesselman, J.D., Eiler, D., Carlson, E.D., Gotrik, M.R., d’Aquino, A.E., Ooms, A.N., Kladwang, W., Carlson, P.D., Shi, X., Costantino, D., Herschlag, D., Lucks, J.B., Jewett, M.C., Kieft, J.S., and Das, R. (2019)
“Computational design of three-dimensional RNA structure and function”
Nature Nanotechnology 14 : 866 – 873
Paper | Link
Koirala, D., Shao, Y., Koldobskaya, Y., Fuller, J.R., Watkins, A.M., Shelke, S.A., Pilipenko, E.V., Das, R., Rice, P.A., and Piccirilli, J.A. (2019) “A conserved RNA structural motif for organizing topology within picornaviral internal ribosome entry sites” Nature Communications 10 : 3629 | Paper | Link
Koodli, R. V., Keep, B., Coppess, K. R., Portela, F., Eterna Participants, and Das, R. (2019) “EternaBrain: automated RNA design through move sets and strategies from an Internet-scale RNA videogame” PLOS Computational Biology 15 (6) : e1007059 | Paper | Link | EternaBrain software
Jarmoskaite, I., Denny, S. K., Vaidyanathan, P. P., Becker, W. R., Andreasson, J. O. L., Layton, C. J., Kappel, K., Shivashankar, V., Sreenivasan, R., Das, R., Greenleaf, W. J., and Herschlag, D. (2019) “A quantitative and predictive model for RNA binding by human Pumilio proteins” Molecular Cell 74 : 966 – 981 | Paper | Link
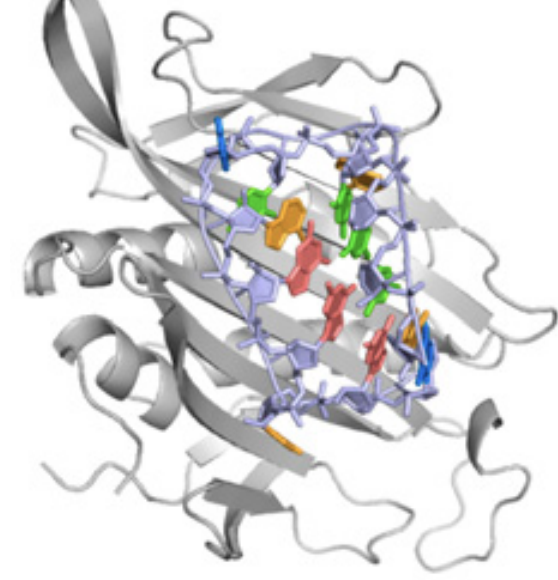 Kappel, K. Jarmoskaite, I. Vaidyanathan, P.P. Greenleaf, W.J. Herschlag, D. and Das R. (2019)
Kappel, K. Jarmoskaite, I. Vaidyanathan, P.P. Greenleaf, W.J. Herschlag, D. and Das R. (2019)
“Blind tests of RNA-protein binding affinity prediction”
PNAS 116 : 8336 – 8341
Paper | Link | Rosetta RNP-ddG Software | ROSIE RNP-ddG Server
Boeynaems, S., Holehouse, A.S., Weinhardt, V., Kovacs, D., Van Lindt, J., Larabell, C., Van Den Bosch, L., Das, R., Tompa, P.S. Pappu, R.V., and Gitler, A.D. (2019) “Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties” PNAS 116 : 7889 – 7898 | Paper | Link
Kappel K. and Das R. (2019) “Sampling native-like structures of RNA-protein complexes through Rosetta folding and docking” Structure 27 (1) : 140 – 151 | Paper | Link | Rosetta RNP modeling software
Koodli, R., Keep, B., Coppess, K.R., Portela, F., Eterna Players, and Das, R. (2019) “RNA design movesets and strategies from an Internet-scale videogame” BioRxiv
2018
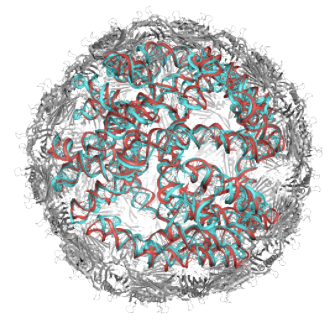 Kappel, K. Liu, S. Larsen, K.P. Skiniotis, G. Puglisi, E.V., Puglisi, J.D., Zhou, Z.H., Zhao, R., and Das, R. (2018)
Kappel, K. Liu, S. Larsen, K.P. Skiniotis, G. Puglisi, E.V., Puglisi, J.D., Zhou, Z.H., Zhao, R., and Das, R. (2018)
“De novo computational RNA modeling into cryoEM maps of large ribonucleoprotein complexes”
Nature Methods 15 : 947 – 954
Paper | Link | DRRAFTER Rosetta Software
Chen, L., Roake, C.M., Freund, A., Batista, P.J., Tian, S., Yin, Y.A., Gajera, C.R., Lin, S., Lee, B., Pech, M.F., Venteicher, A.S., Das, R., Chang, H.Y., and Artandi, S.E. (2018) “An activity switch in human telomerase based on RNA conformation and shaped by TCAB1” Cell 174 (1) : 218-230 | Paper | Link
Shi, J., Eterna players, Das, R., and Pande, V.S. (2018) “SentRNA: Improving computational RNA design by incorporating a prior of human design strategies” BioRxiv | Preprint | SentRNA Software
![]() Watkins, A.M., Geniesse, C., Kladwang, W., Zakrevsky, P., Jaeger, L., and Das, R. (2018)
Watkins, A.M., Geniesse, C., Kladwang, W., Zakrevsky, P., Jaeger, L., and Das, R. (2018)
“Blind prediction of noncanonical RNA structure at atomic accuracy”
Science Advances 4 (5) : eaar5316
Paper | Link | Stanford Digital Repository PURL | ROSIE server
Wu, M.J., Andreasson, J.O.L, Kladwang, W., Greenleaf, W.J., Eterna players, and Das, R. (2018) “Prospects for recurrent neural network models to learn RNA biophysics from high-throughput data” BioRxiv | Preprint
Das, R. (2018) “LIkelihood-based Fits of Folding Transitions (LIFFT) for Biomolecule Mapping Data” BioRxiv | Preprint | LIFFT Software
Gracia, B., Al-Hashimi, H.M., Bisaria N., Das, R., Herschlag, D., and Russell, R. (2018) “Hidden structural modules in a cooperative RNA folding transition” Cell Reports 22 : 3240 – 3250 | Link
Moretti, R., Lyskov, S., Das, R., Meiler, J., and Gray, J.J. (2018) “Web-accessible molecular modeling with Rosetta: The Rosetta Online Server That Includes Everyone (ROSIE)” Protein Science 27 (1) : 259 – 268 | Paper | Link | ROSIE Servers
2017
Alford, R.F., Leaver-Fay, A., Jeliazkov, J.R., O’Meara, M.J., DiMaio, F.D., Park, H., Shapovalov, M.V., Renfrew, P.D., Mulligan, V.K., Kappel, K., Labonte, J.W., Pacella , M.S., Bonneau, R., Bradley, P., Dunbrack, Jr., R.L., Das, R., Baker, D., Kuhlman, B., Kortemme, T., and Gray, J.J. (2017) “The Rosetta all-atom energy function for macromolecular modeling and design” Journal of Chemical Theory and Computation 13 (6) : 3031 – 3048 | Paper | Link
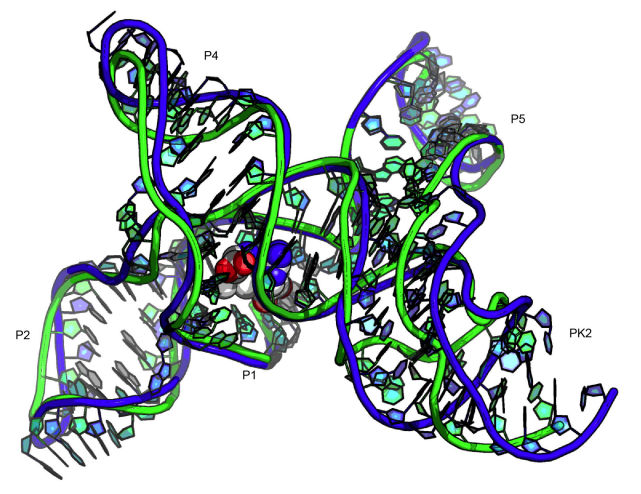 Miao, Z., Adamiak, R. W., Antczak, M., Batey, R. T., Becka, A. J., Biesiada, M., Boniecki, M. J., Bujnicki, J. M., Chen, S.-J., Cheng, C. Y., Chou, F.-C., Ferré-D’Amaré, A. R., Das, R., Dawson, W. K., Ding, F., Dokholyan, N. V., Dunin-Horkawicz, S., Geniesse, C., Kappel, K., Kladwang, W., Krokhotin, A., Lach, G. E., Major, F., Mann, T. H., Magnus, M., Pachulska-Wieczorek, K., Patel, D. J., Piccirilli, J. A., Popenda, M., Purzycka, K. J., Ren, A., Rice, G. M., Santalucia, J. Jr., Sarzynska, J., Szachniuk, M., Tandon, A., Trausch, J. J., Tian, S., Wang, J., Weeks, K. M., Williams, B. II, Xiao, Y., Xu, X., Zhang, D., Zok, T., and Westhof, E. (2017)
Miao, Z., Adamiak, R. W., Antczak, M., Batey, R. T., Becka, A. J., Biesiada, M., Boniecki, M. J., Bujnicki, J. M., Chen, S.-J., Cheng, C. Y., Chou, F.-C., Ferré-D’Amaré, A. R., Das, R., Dawson, W. K., Ding, F., Dokholyan, N. V., Dunin-Horkawicz, S., Geniesse, C., Kappel, K., Kladwang, W., Krokhotin, A., Lach, G. E., Major, F., Mann, T. H., Magnus, M., Pachulska-Wieczorek, K., Patel, D. J., Piccirilli, J. A., Popenda, M., Purzycka, K. J., Ren, A., Rice, G. M., Santalucia, J. Jr., Sarzynska, J., Szachniuk, M., Tandon, A., Trausch, J. J., Tian, S., Wang, J., Weeks, K. M., Williams, B. II, Xiao, Y., Xu, X., Zhang, D., Zok, T., and Westhof, E. (2017)
“RNA-Puzzles Round III: 3D RNA structure prediction of five riboswitches and one ribozyme”
RNA 23 (5) : 655 – 672
Paper | Link
2016
2015
 Miao, Z., Adamiak, R.W., Blanchet, M-F., Boniecki, M., Bujnicki, J.M., Chen, S-J., Cheng, C.Y., Chojnowski, G., Chou, F-C., Cordero, P., Cruz, J.A., Ferré-D’Amaré, A, Das, R., Ding, F., Dokholyan, N.V., Dunin-Horkawicz, S., Kladwang, W., Krokhotin, A., Lach, G., Magnus, M., Major, F., Mann, T.H., Masquida, B., Matelska, D., Meyer, M., Peselis, A., Popenda, M., Purzycka, K.J., Serganov, A., Stasiewicz, J., Szachniuk, M., Tandon, A., Tian, S., Wang, J., Xiao, Y., Xu, X., Zhang, J, Zhao, P., Zok, T., and Westhof, E. (2015)
Miao, Z., Adamiak, R.W., Blanchet, M-F., Boniecki, M., Bujnicki, J.M., Chen, S-J., Cheng, C.Y., Chojnowski, G., Chou, F-C., Cordero, P., Cruz, J.A., Ferré-D’Amaré, A, Das, R., Ding, F., Dokholyan, N.V., Dunin-Horkawicz, S., Kladwang, W., Krokhotin, A., Lach, G., Magnus, M., Major, F., Mann, T.H., Masquida, B., Matelska, D., Meyer, M., Peselis, A., Popenda, M., Purzycka, K.J., Serganov, A., Stasiewicz, J., Szachniuk, M., Tandon, A., Tian, S., Wang, J., Xiao, Y., Xu, X., Zhang, J, Zhao, P., Zok, T., and Westhof, E. (2015)
“RNA-Puzzles Round II: Assessment of RNA structure prediction programs applied to three large RNA structures.”
RNA 21 (6) : 1066 – 1084
Paper | Link
2014
Lipfert, J., Skinner, G.M., Keegstra, J.M., Hensgens, T., Jager, T., Dulin, D., Köber, M., Yu, Z., Donkers, S.P., Das, R., and Dekker, N.H. (2014) “Double-stranded RNA under force and torque: Similarities to and striking differences from double-stranded DNA.” Proceedings of the National Academy of Sciences U.S.A. 111 (43) : 15408 – 15413 | Paper | Link
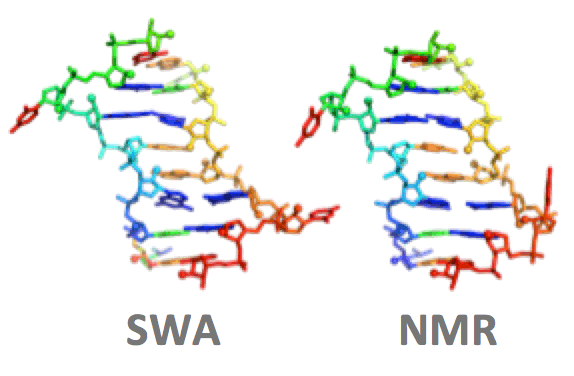 Sripakdeevong, P., Cevec, M., Chang, A.T., Erat, M.C., Ziegeler, M., Zhao, Q., Fox, G.E., Gao, X., Kennedy, S.D., Kierzek, R., Nikonowicz, E.P., Schwalbe, H., Sigel, R.K.O., Turner, D.H., and Das, R. (2014)
Sripakdeevong, P., Cevec, M., Chang, A.T., Erat, M.C., Ziegeler, M., Zhao, Q., Fox, G.E., Gao, X., Kennedy, S.D., Kierzek, R., Nikonowicz, E.P., Schwalbe, H., Sigel, R.K.O., Turner, D.H., and Das, R. (2014)
“Consistent structure determination of noncanonical RNA motifs from 1H NMR chemical shift data”
Nature Methods 11 : 413 – 416
Paper | Link | Server
Kryshtafovych, A., Moult, J., Bales, P., Bazan, J.F., Burgin, A., Chen, C., Cochran, F.V., Craig, T.K., Das, R., Fass, D., Garcia-Doval, C., Herzberg, O., Lorimer, D., Luecke, H., Ma, X., Nelson, D., van Raaij, M.J., Rohwer, F., Segall, A., Seguritan, V., Zeth, K., and Schwede, T. (2014) “Challenging the state-of-the-art in protein structure prediction: Highlights of experimental target structures for the 10th critical assessment of techniques for protein structure prediction experiment CASP10” Proteins 82 (S2) : 26 – 42 | Paper | Link
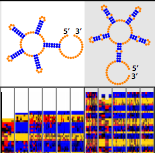 Lee, J., Kladwang, W., Lee, M., Cantu, D., Azizyan, M., Kim, H., Limpaecher, A., Yoon, S., Treuille, A., Das, R., and EteRNA Participants (2014)
Lee, J., Kladwang, W., Lee, M., Cantu, D., Azizyan, M., Kim, H., Limpaecher, A., Yoon, S., Treuille, A., Das, R., and EteRNA Participants (2014)
“RNA design rules from a massive open laboratory”
Proceedings of the National Academy of Sciences U.S.A. 111 (6) : 2122 – 2127
Paper | Link | SHAPE Data Files | EternaBot Software
2013
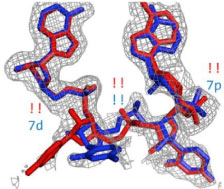 Chou, F.-C., Sripakdeevong, P., Dibrov, S.M, Hermann, T., and Das, R. (2013)
Chou, F.-C., Sripakdeevong, P., Dibrov, S.M, Hermann, T., and Das, R. (2013)
“Correcting pervasive errors in RNA crystallography through enumerative structure prediction”
Nature Methods 10 : 74 – 76
Paper | Link | ERRASER Server
Drew, K., Renfrew, P.D., Craven, T., Butterfoss, G.L., Chou, F.-C., Lyskov, S., Bullock, B.N., Watkins, A., Leaver-Fay, A., Kuhlman, B., Gray, J.J., Bradley, P., Kirshenbaum, K., Arora, P.S., Das, R., and Bonneau, R. (2013) “Adding diverse noncanonical backbones to Rosetta: enabling peptidomimetic and foldamer design” PLOS One 8 (7) : e67051 | Paper | Link
Lyskov, S., Chou, F.-C., Ó Conchúir, S., Der, B.S., Drew, K., Kuroda, D., Xu, J., Weitzner, B.D., Renfrew, P.D., Sripakdeevong, P., Borgo, B., Havranek, J.J., Kuhlman, B., Kortemme, T., Bonneau, R., Gray, J.J., and Das, R. (2013) “Serverification of molecular modeling applications: the Rosetta online server that includes everyone (ROSIE)” PLOS One 8 (5) : e63906 | Paper | Link | Server
Adams, P.D., Baker, D., Brunger, A.T., Das, R., DiMaio, F., Read, R.J., Richardson, D.C., Richardson, J.S., and Terwilliger, T.C. (2013) “Advances, interactions, and future developments in the CNS, Phenix, and Rosetta structural biology software systems” Annual Review of Biophysics 42 : 265 – 287 | Paper | Link
Molski, M.A., Goodman, J.L, Chou, F-C., Baker, D., Das, R., and Schepartz, A. (2013) “Remodeling a beta-peptide bundle” Chemical Science 4 : 319 – 324 | Paper | Link | Design Server
2012
 Cruz, J.A., Blanchet, M., Boniecki, M., Bujnicki, J., Chen, S-J., Cao, S., Das, R., Ding, F., Dokholyan, N.V., Flores, S.C., Huang, L., Lavender, C.A., Lisi, V., Major, F., Mikolajczak, K., Patel, D.J., Philips, A., Puton, T., Santalucia, J., Sijenyi, F., Hermann, T., Rother, K., Rother, M., Serganov, A., Skorupski, M., Soltysinski, T., Sripakdeevong, P., Tuszynska, I., Weeks, K.M., Waldsich, C., Wildauer, M., Leontis, N.B., and Westhof, E. (2012)
Cruz, J.A., Blanchet, M., Boniecki, M., Bujnicki, J., Chen, S-J., Cao, S., Das, R., Ding, F., Dokholyan, N.V., Flores, S.C., Huang, L., Lavender, C.A., Lisi, V., Major, F., Mikolajczak, K., Patel, D.J., Philips, A., Puton, T., Santalucia, J., Sijenyi, F., Hermann, T., Rother, K., Rother, M., Serganov, A., Skorupski, M., Soltysinski, T., Sripakdeevong, P., Tuszynska, I., Weeks, K.M., Waldsich, C., Wildauer, M., Leontis, N.B., and Westhof, E. (2012)
“RNA-Puzzles: A CASP-like evaluation of RNA three-dimensional structure prediction”
RNA 18 (4) : 610 – 625
Paper | Link
2011
Rocca-Serra, P., Bellaousov, S., Birmingham, A., Chen, C., Cordero, P., Das, R., Davis-Neulander, L., Duncan, C.D., Halvorsen, M., Knight, R., Leontis, N.B., Mathews, D.H., Ritz, J., Stombaugh, J., Weeks, K.M., Zirbel, C.L., and Laederach, A. (2011) “Sharing and archiving nucleic acid structure mapping data” RNA 17 (7) : 1204 – 1212 | Paper | Link
Leaver-Fay, A., Tyka, M., Lewis, S.M., Lange, O.F., Thompson, J. Jacak, R., Kaufmann, K.W., Renfrew, P.D., Smith, C.A., Sheffler, W., Davis, I.W., Cooper, S., Treuille, A., Mandell, D.J., Richter, F., Ban, YE., Fleishman, S.J., Corn, J.E., Kim, D.E., Lyskov, S., Berrondo, M., Mentzer, S., Popovic, Z., Havranek, J.J., Karanicolas, J., Das, R., Meiler, J., Kortemme, T., Gray, J.J., Kuhlman, B., Baker, D., and Bradley, P. (2011) “ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules” Methods in Enzymology 487 : 545 – 574 | Paper | Link
2010
2009
Raman, S., Vernon, R., Thompson, J., Tyka, M., Sadreyev, R., Pei, J., Kim, D., Kellogg, E., DiMaio, F., Lange, O., Kinch, L., Sheffler, W., Kim, B.H., Das, R., Grishin, N.V., and Baker, D. (2009) “Structure prediction for CASP8 with all-atom refinement using Rosetta.” Proteins 77 (S9) : 88 – 99 | Paper | Link
Schwede T., Sali, A., Honig, B., Levitt, M., Berman, H.M., Jones, D., Brenner, S.E., Burley, S.K., Das, R., Dokholyan, N.V., Dunbrack, R.L., Fidelis, K., Fiser, A., Godzik, A., Huang, Y.J., Humblet, C., Jacobson, M.P., Joachimiak, A., Krystek, S.R., Kortemme, T., Kryshtafovych, A., Montelione, G.T., Moult, J., Murray, D., Sanchez, R., Sosnick, T.R., Standley, D.M., Stouch, T., Vajda, S., Vasquez, M., Westbrook, J.D., and Wilson, I.A. (2009) “Outcome of a workshop on applications of protein models in biomedical research” Structure 17 (2) : 151 – 159 | Paper | Link
2008
Das, R., Kudaravalli, M., Jonikas, M., Laederach, A., Fong, R., Schwans, J.P., Baker, D., Piccirilli, J.A., Altman, R.B., and Herschlag, D. (2008) “Structural inference of native and partially folded RNA by high throughput contact mapping” Proceedings of the National Academy of Sciences U.S.A. 105 (11) : 4144 – 4149 | Paper | Link | Scripts
2007
Das, R., Qian, B., Raman, V.S., Vernon, R., Thompson, J., Bradley, P., Khare, S., Tyka, M., Bhat, D., Chivian, D.C., Kim, D.E., Sheffler, W., Malmstrom, L., Wollacott, A., Wang, C., Andre, I., and Baker, D. (2007) “Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home” Proteins 69 (S8) : 118 – 128 | Paper | Link
Blum, B., Jordan, M.I., Kim, D., Das, R., Bradley, P., and Baker, D. (2007) “Feature selection methods for improving protein structure prediction with Rosetta” in Advances in Neural Information Processing Systems (NIPS) 21, eds. Platt, J., Koller, D., Singer, Y., and McCallum, A., MIT Press. | Paper | Link
2006
2005
2004
2003
Das, R., Kwok, L.W., Millet, I.S., Bai, Y., Mills, T.T., Jacob, J., Maskel, G.S., Seifert, S., Simon, M.G.J., Thiyagarajan, P., Doniach, S., Pollack, L., and Herschlag, D. (2003) “The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme.” Journal of Molecular Biology 332 (2) : 311 – 319 | Paper | Link
Saunders, R., Kneissl, R., Grainge, K., Grainger, W.F., Jones, M.E., Maggi, A., Das, R., Edge, A.C., Lasenby, A.N., Pooley, G.G., Shigeru, J.M., Tsuruta, T., Yamashita, K., Tawara, Y., Furuzawa, A., Harada, A., and Hatsukade, I. (2003) “A measurement of H0 from Ryle Telescope, ASCA and ROSAT observations of Abell 773.” Monthly Notices of the Royal Astronomical Society 341 (3) : 937 – 940 | Paper | Link
2002
Grainger, W.F., Das, R., Grainge, K., Jones, M. E., Kneissl, R., Pooley, G.G., and Saunders, R.D.E. (2002) “A maximum-likelihood approach to removing radio sources from observations of the Sunyaev-Zel’dovich effect, with application to Abell 611.” Monthly Notices of the Royal Astronomical Society 337 (4) : 1207 – 1214 | Paper | Link
Cotter G., Buttery, H.J., Rawlings, S., Croft, S., Hill, G.J., Gay, P., Das, R., Drory, N., Grainge, K., Grainger, W.F., Jones, M.E., Pooley, G.G., and Saunders, R. (2002) “Detection of a cosmic microwave background decrement towards a cluster of mJy radio sources.” Monthly Notices of the Royal Astronomical Society 331 (1) : 1 – 6 | Paper | Link
2001
2000
Das, R. (2000) “Theoretical Basis of Likelihood Methods in Molecular Phylogenetic Inference” M. Res. Thesis, Dept. of Biology, University College London. Supervisors: Z. Yang & J. Mallet. | Paper
1999
Das, R. (1999) “Analysis of Sunyaev-Zel’dovich Decrements” M. Phil. Thesis, Dept. of Physics, Cambridge University. Supervisor: R. Saunders. | Paper

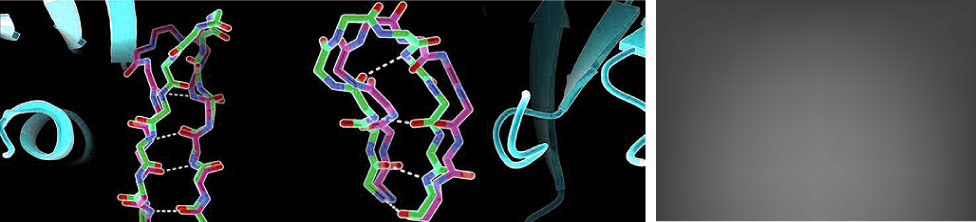

 Kretsch, R.C., Xu, L., Zheludev, I.N., Zhou, X., Huang, R., Nye, G., Li, S., Zhang, K., Chiu, W., Das, R. (2024)
Kretsch, R.C., Xu, L., Zheludev, I.N., Zhou, X., Huang, R., Nye, G., Li, S., Zhang, K., Chiu, W., Das, R. (2024) Das, R., Kretsch, R., Simpkin, A., Mulvaney, T., Pham, P., Rangan, R., Bu, F., Keegan, R., Topf, M., Rigden, D., Miao, Z., Westhof, E. (2023)
Das, R., Kretsch, R., Simpkin, A., Mulvaney, T., Pham, P., Rangan, R., Bu, F., Keegan, R., Topf, M., Rigden, D., Miao, Z., Westhof, E. (2023)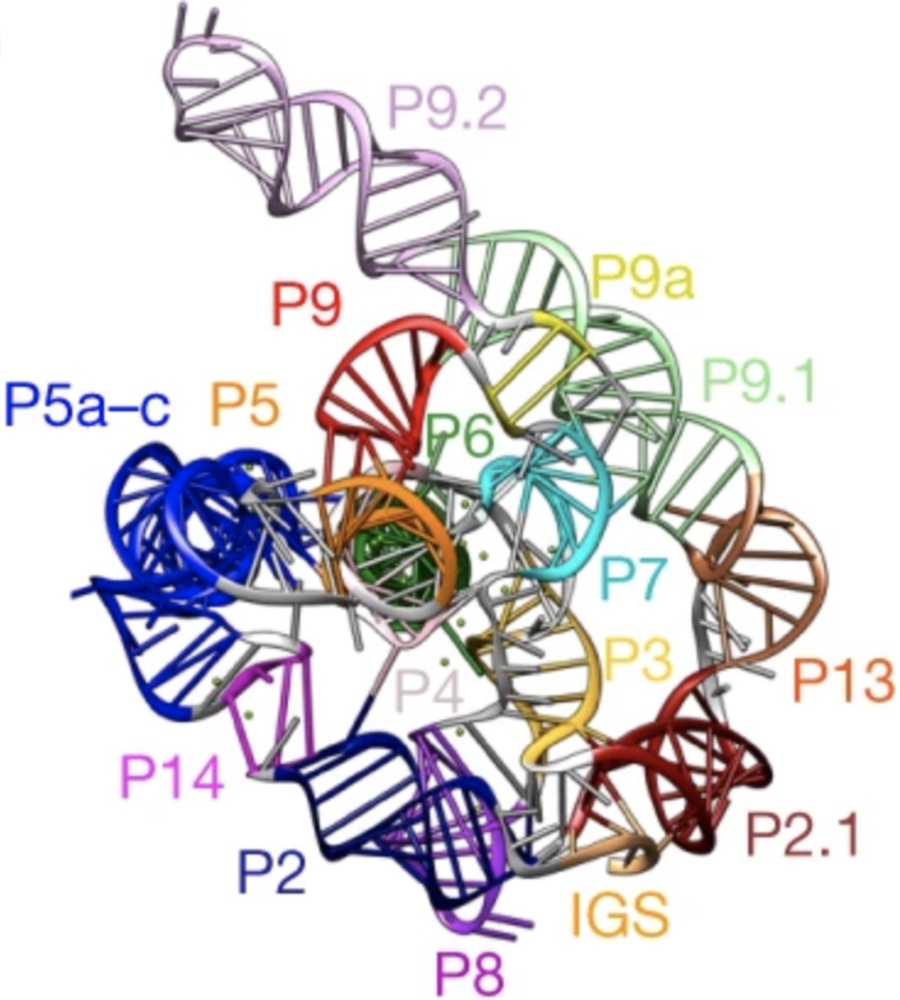 Su, Z., Zhang, K., Kappel, K., Li, S., Palo, M.Z., Pintilie, G.D., Rangan, R., Lou, B., Wei, Yuquan., Das, R., Chiu, W. (2021)
Su, Z., Zhang, K., Kappel, K., Li, S., Palo, M.Z., Pintilie, G.D., Rangan, R., Lou, B., Wei, Yuquan., Das, R., Chiu, W. (2021) Kappel, K., Zhang, K., Su, Z., Watkins, A.M., Kladwang, W., Li, S., Pintilie, G., Topkar, V.V., Rangan, R., Zheludev, I.N., Yesselman, J.D., Chiu, W., and Das, R. (2020)
Kappel, K., Zhang, K., Su, Z., Watkins, A.M., Kladwang, W., Li, S., Pintilie, G., Topkar, V.V., Rangan, R., Zheludev, I.N., Yesselman, J.D., Chiu, W., and Das, R. (2020) Zhang, K., Li, S., Kappel, K., Pintilie, G., Su, Z., Mou, T.-C., Schmid, M. F., Das, R., Chiu, W. (2019)
Zhang, K., Li, S., Kappel, K., Pintilie, G., Su, Z., Mou, T.-C., Schmid, M. F., Das, R., Chiu, W. (2019) Tian, S., Kladwang, W., and Das, R. (2018)
Tian, S., Kladwang, W., and Das, R. (2018) Cheng, C.Y., Kladwang, W., Yesselman, J.D., and Das, R. (2017)
Cheng, C.Y., Kladwang, W., Yesselman, J.D., and Das, R. (2017)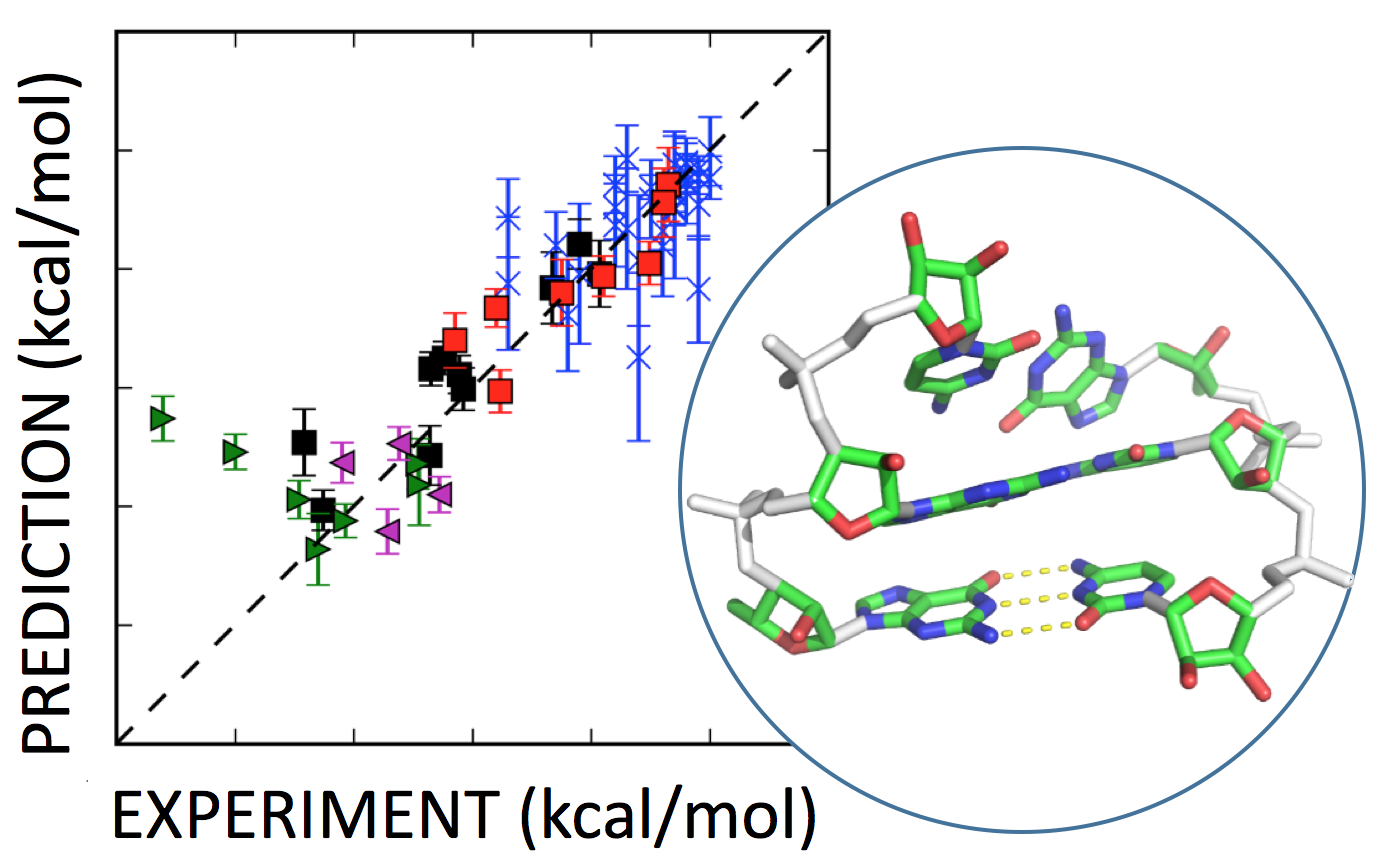 Chou, F-C., Kladwang, W., Kappel, K., and Das, R. (2016)
Chou, F-C., Kladwang, W., Kappel, K., and Das, R. (2016) Anderson-Lee, J., Fisker, E., Kosaraju,V., Wu, M., Kong, J., Lee, J., Lee, M., Zada, M., Treuille, A., Das, R., and Eterna Players (2016)
Anderson-Lee, J., Fisker, E., Kosaraju,V., Wu, M., Kong, J., Lee, J., Lee, M., Zada, M., Treuille, A., Das, R., and Eterna Players (2016) Cheng, C.Y., Chou, F.-C., Kladwang, W., Tian, S., Cordero, P., and Das, R. (2015)
Cheng, C.Y., Chou, F.-C., Kladwang, W., Tian, S., Cordero, P., and Das, R. (2015) Treuille, A., and Das, R. (2014)
Treuille, A., and Das, R. (2014) Tian, S., Cordero, P., Kladwang, W., and Das, R. (2014)
Tian, S., Cordero, P., Kladwang, W., and Das, R. (2014) Das, R. (2013)
Das, R. (2013) Bida, J.P., and Das, R. (2012)
Bida, J.P., and Das, R. (2012) Sripakdeevong, P., Kladwang, W., and Das, R. (2012)
Sripakdeevong, P., Kladwang, W., and Das, R. (2012) Kladwang, W., VanLang, C.C., Cordero P., and Das, R. (2011)
Kladwang, W., VanLang, C.C., Cordero P., and Das, R. (2011) Kladwang, W., and Das, R. (2010)
Kladwang, W., and Das, R. (2010)